Abstract
Vitis amurensis ‘Shuanghong’ is a hybrid offspring of wild grapes. This study first releases the complete chloroplast genome of V. amurensis ‘Shuanghong’ and subjected the sample to phlogenetic analysis. The chloroplast genome is 161,558 bp in length, and comprises a small single-copy region (19,336 bp) and a large single-copy region (89,744 bp), which are seperated by a pair of inverted repeat regions. The chloroplast genome encodes 133 genes, including 88 CDSs, 8 rRNA genes, and 37 tRNA genes. The phylogenetic tree showed that V. amurensis ‘Shuanghong’ is most closely related to Vitis vinifera.
Vitis amurensis ‘Shuanghong’ shows extremely strong cold-resistant and downy mildew resistance properties. So, it is widely used for breeding of grapes against downy mildew (Zhao Y et al. Citation2020). It is also an excellent wine brewing variety because of high anthocyanins content (Zhao Q et al. Citation2016). The complete chloroplast genome of V. amurensis ‘Shuanghong’ was assembled (GenBank; MT479164) and subject to phlogenetic analysis. It offers useful information for the resistance breeding of grapevine.
Genomic DNA was extracted from leaves of V. amurensis ‘Shuanghong’ grown at the Modern Engineering Training Center (31°11′N, 121°29′W) and stored at the Center for Viticulture and Enology, Shanghai Jiao Tong University, number was “Shuanghong”. This DNA was used for the preparation of a 400 bp small-fragment DNA library and then sequenced using the HiSeq PE150 sequencing platform (Illumina, CA, USA). A total of 3.22 Gb clean reads were obtained. The complete chloroplast genome was assembled by A5-MiSeqv20150522 (Coil et al. Citation2015) and SPAdesv3.9.0 (Bankevich et al. Citation2012) software. The chloroplast genome of V. vinifera (GenBank; DQ424856) was used as a reference (Jansen et al. Citation2006) and the genome annotation also referred to the chloroplast genome of V. vinifera.
The V. amurensis ‘Shuanghong’ chloroplast genome is 161,558 bp in length, including two inverted repeat regions (26,239 bp each) that are separated by a small single-copy region (19,336 bp) and a large single-copy region (89,744 bp). The chloroplast genome contains 133 single genes, including 88 protein-coding genes (CDS), 8 rRNA, and 37 tRNA genes. The GC content and AT content of the grape genome is 36.58% and 63.42%, respectively. Among these genes, the majority are single copy, whereas 8 CDS (rp12, rp123, rps7, rps12, rps19, ycf2, ycf15, ndhB), 7 tRNAs (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, trnN-GUU) and 4 rRNAs (rrn4.5, rrn5, rrn-16, rrn-23) occur as double copies.
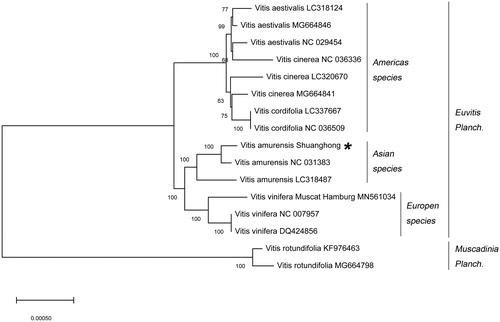
A neighbor-joining (NJ) phylogenetic tree was constructed using 16 Vitis species through the MEGA X (Kumar et al. Citation2018). To identify the phylogenetic position of V. amurensis ‘Shuanghong’ within the family Vitaceae, the phylogenetic tree showed that the 16 Vitis species are clustered into two orders (). The V. amurensis ‘Shuanghong’ was phylogenetically closer to Vitis vinifera of Europen species than species in other generas.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in GenBank: MT479164 at https://www.ncbi.nlm.nih.gov/genbank/.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Son P, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- Jansen RK, Kaittanis C, Saski C, Lee S-B, Tomkins J, Alverson AJ, Daniell H. 2006. Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol. 6(1):32.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Zhao Q, He F, Reeves MJ, Pan Q-H, Duan C-Q, Wang J. 2016. Expression of structural genes related to anthocyanin biosynthesis of Vitis amurensis. J Res. 27(3):647–657.
- Zhao Y, Wang Z-X, Yang Y-M, Liu H-S, Shi G-L, Ai J. 2020. Analysis of the cold tolerance and physiological response differences of amur grape (Vitis amurensis) germplasms during overwintering. Sci Hortic. 259:108760.