Abstract
The complete mitochondrial genome was sequenced in Levanidov’s charr Salvelinus levanidovi. The genome sequences are 16,624 bp, and the gene arrangement, composition, and size are similar to the charr genomes. The level of divergence between S. levanidovi and charr belonging to the genus Salvelinus was in the range from 4.80% to 3.65%. Molecular phylogeny provides new evidence that S. levanidovi is closely related to the common ancestor of the genus Salvelinus. The present study confirms that S. fontinalis, S. levanidovi, S. leucomaensis, and S. namaycus form a basal group of taxa, each of them belongs to an independent evolutionary line.
The Levanidov’s charr Salvelinus levanidovi was first described as an endemic species inhabiting a small range in rivers of the northern basin of the Sea of Okhotsk (Chereshnev et al. Citation1989). According to comparative data, S. levanidovi is characterized by the prevalence of plesiomorphic morphological characters (Chereshnev et al. Citation2002). In addition, S. levanidovi has preserved a primitive karyotype, which is also typical of S. namaycush and S. fontinalis (Frolov and Frolova Citation2004), and the ancestral mitochondrial genome (Oleinik et al. Citation2003). Molecular phylogenies, inferred from different DNA markers using various methods of analyses, indicate that basal position within the genus could be occupied by any of the following four species: S. fontinalis, S. levanidovi, S. leucomaensis, or S. namaycus (e.g. Oleinik, Skurikhina, et al. Citation2015; Osinov, Senchukova, et al. Citation2015, and references therein). Therefore, to research higher-level relationships among Salvelinus and to find out which of the charr species is the ‘most basal clade’, it is important to obtain the complete mitochondrial genome of S. levanidovi.
We sequenced and described the complete mitogenome of S. levanidovi for the first time in this study. Salvelinus levanidovi were collected in the middle flow of the Yama River (Sea of Okhotsk basin, Russia; 59°41′N, 154°21′E), a type locality of Levanidov’s charr (Chereshnev et al. Citation1989). The fish specimen is stored in the collection of the Genetics Laboratory, NSCMB FEB RAS, Vladivostok, Russia (www.imb.dvo.ru) with accession number LVYA94.030. Totally 5 pairs of primers were used, which were designed based on public sequences available in the GenBank for salmonid fish. The sequenced fragments were assembled into a complete mitogenome and annotated by comparing with published mitogenomes of charr using Geneious R11 (http://www.geneious.com/).
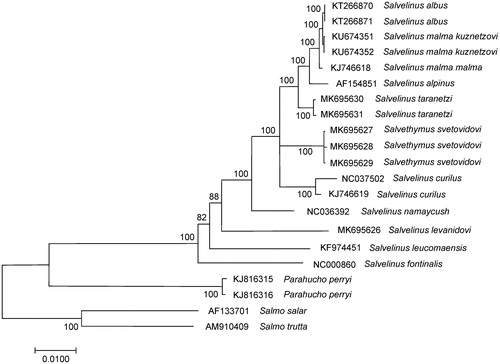
The complete mitogenome of native S. levanidovi was 16,624 bp in length. Like charr mitogenomes (Balakirev et al. Citation2016; Oleinik et al. Citation2019), the overall base composition was 28.1% of A, 26.4% of T, 28.6% of C, and 16.9% of G with a slight A + T bias (54.5%). The comparison of complete mitogenome was obtained with 20 mitogenomes of related groups available in the GenBank, including genera Salvelinus, Parahucho, and Salmo point to the relationships of S. levanidovi to charr species (). Salvelinus levanidovi was phylogenetically positioned with other charr, showing a clear divergence from them. The average level of total sequence divergence (Dxy) between them was 0.0390 ± 0.001; these values correspond to the level of interspecific variability in the genus (Oleinik, Skurikhina, et al. Citation2015; Osinov, Senchukova, et al. Citation2015). The highest divergence was found between the mitogenomes of S. levanidovi and S. fontinalis (0.0480 ± 0.0016) as well of S. levanidovi and S. leucomaenis (0.0423 ± 0.0016). At the same time, our S. levanidovi specimen showed similar sequence divergence (0.0365 ± 0.0015 on average) from other charrs in GenBank, including charrs from East Asia to North American. Вased on the variability of mitogenomes, S. fontinalis, S. levanidovi, S. leucomaensis, and S. namaycus form a basal group of taxa, each of them belonging to an independent evolutionary line. The present study confirms that S. levanidovi is closely related to the common ancestor of the genus Salvelinus.
Figure 1. Maximum likelihood (ML) tree constructed based on the comparison of complete mitochondrial genome sequences of S. levanidovi and other GenBank representatives of the family Salmonidae. The tree is based on the GTR plus gamma plus invariant sites (GTR + G + I) model of nucleotide substitution. Genbank accession numbers for all sequences are listed in the figure. Numbers at the nodes indicate bootstrap probabilities from 1000 replications (values below 80% are omitted).
Acknowledgements
We are grateful to Dr. S.V. Frolov (NSCMB FEB RAS, Vladivostok, Russia) for the Salvelinus levanidovi specimens. The research on mitochondrial genome sequencing was conducted at the School of Natural Sciences, Far Eastern Federal University, Vladivostok, Russia. The data analysis and manuscript preparation were conducted at the A.V. Zhirmunsky National Scientific Center of Marine Biology, Vladivostok, Russia.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the National Center for Biotechnology Information database (NCBI/GenBank) at https://https.ncbi.nlm.nih.gov/, reference number MK695626.
Additional information
Funding
References
- Balakirev ES, Parensky VA, Kovalev MYu, Ayala FJ. 2016. Complete mitochondrial genome of the stone char Salvelinus kuznetzovi (Salmoniformes, Salmonidae). Mitochondrial DNA (Part B). 1(1):287–288.
- Chereshnev IA, Skopets MB, Gudkov PK. 1989. A new charr species, Salvelinus levanidovi sp. nov., from the basin of the sea of Okhotsk. Vopr Ikhtiol. 29(5):691–704.
- Chereshnev IA, Volobuev VV, Shestakov AV, Frolov SV. 2002. Salmonoid fishes in Russian north-east. Vladivostok: Dalnauka.
- Frolov SV, Frolova VN. 2004. Karyological differentiation of northern Dolly Varden and sympatric chars of the genus Salvelinus in northeastern Russia. Environ Biol Fish. 69(1–4):441–447.
- Oleinik AG, Skurikhina LA, Brykov VA. 2003. Genetic differentiation of three sympatric charr species from the genus Salvelinus inferred from PCR–RFLP analysis of mitochondrial DNA. Russ J Genet. 39(8):924–929.
- Oleinik AG, Skurikhina LA, Brykov VA. 2015. Phylogeny of charrs of the genus Salvelinus based on mitochondrial DNA data. Russ J Genet. 51(1):55–68.
- Oleinik AG, Skurikhina LA, Kukhlevsky AD, Semenchenko AA. 2019. First report of three complete mitochondrial genomes of the long-finned charr Salvethymus svetovidovi Chereshnev et Skopetz, 1990 (Salmoniformes: Salmonidae) with phylogenetic consideration. Mitochondrial DNA Part B. 4(2):2464–2466.
- Osinov AG, Senchukova AL, Mugue NS, Pavlov SD, Chereshnev IA. 2015. Speciation and genetic divergence of three species of charrs from ancient Lake El’gygytgyn (Chukotka) and their phylogenetic relationships with other representatives of the genus Salvelinus. Biol J Linn Soc. 116(1):63–85.
