Abstract
Cervus canadensis (Erxleben, 1777) has been used as a model species of Chronic Wasting Disease (CWD). We completed the mitochondrial genome of C. canadensis, susceptible to the CWD. Its length is 16,428 bp, identical to the previous mitochondrial genome of C. canadensis nannodes, and 37 genes (13 protein-coding genes, two rRNAs, and 22 tRNAs) were identified. It may reflect the extreme decrease of tule elk population in 1870s and CWD is not related to genetic elements on mitochondrial genome. Phylogenetic trees show that our mitochondrial genome is clustered with the previously sequenced mitochondrial genome of C. canadensis nannodes .
Chronic wasting disease (CWD), a fatal neurological prion disease accompanying abnormal behavior, was recognized in 1967 in captive mule deer (Odocoileus hemionus) and hybrids of mule deer and white tailed deer (Odocoileus hemionus virginianus) in wildlife facilities in Colorado, USA (Williams and Young Citation1980). Now, it has been identified in North America, Canada, Republic of Korea, Norway, and Finland (Sohn et al. Citation2002; Kim et al. Citation2005; Williams Citation2005; Benestad et al. Citation2016). To understand the precise molecular-level mechanism of CWD, elks (Cervus canadensis ) were used as a model species. Its genetic background will be useful to undedrstand in various aspects including their maternal lineage. Some of them displayed susceptibility to CWD and some were resistance. Mitogenome of the resistance tule elk was successfully sequenced (doi:10.1080/23802359.2020.1772689), which can be used a reference mitochondrial genome to evaluate their genetic diversity before dissecting molecular mechanisms of this disease using them. Meanwhile, mitochondrial genome of the susceptible tule elk individual to CWD selected to investigate the genes involved sensitivity of the CWD was sequenced to know their genetic diversity.
The brain tissue of C. canadensis nannodes collected in Namhae-gun, Gyeongsangnamdo, Korea (Voucher in Animal and Plant Quarantine Agency, Korea; APQA-R-19FC017; 36.125833 N, 128.199515E) was used for extracting DNA with DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeqX at Macrogen Inc. Mitochondrial genome was assembled by Velvet 1.2.10 (Zerbino and Birney Citation2008) and gap sequences was filled with SOAPGapCloser 1.12 (Zhao et al. Citation2011). Base pairs on its mitochondrial genome were confirmed using BWA 0.7.17 (Li Citation2013) and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for annotation based on C. canadensis nannodes mitochondrial genome (MT430939; Kim et al. Citation2020).
The mitochondrial genome of C. canadensis (GenBank accession is MT534583) is 16,428 bp and contains 37 genes (13 protein-coding genes, two rRNAs, and 22 tRNAs) and GC ratio is 38.0%. It is identical to the first mitochondrial genome of C. cnadensis nannodes (MT430939; Kim et al. Citation2020), which is similar to the cases of insect mitochondrial genome of Laodelphax striatellus (one SNP) (Seo et al. Citation2019) as well as chloroplast genomes of Salix koriyanagi (Park et al. Citation2019), Populus alba × Populus glandulosa (Park, Kim, Xi, et al. Citation2019), and Coffea arabica (Park et al. Citation2020). It indicates that susceptibility of the CWD may not related to mitochondrial genomic elements. In addition, tule elks which are susceptible and resistible to the CWD share with the same material genetic background because of both have the identical mitochondrial genome. Moreover, it can also be one of evidences of bottle effect occurred in 1870s (McCullough et al. Citation1996).
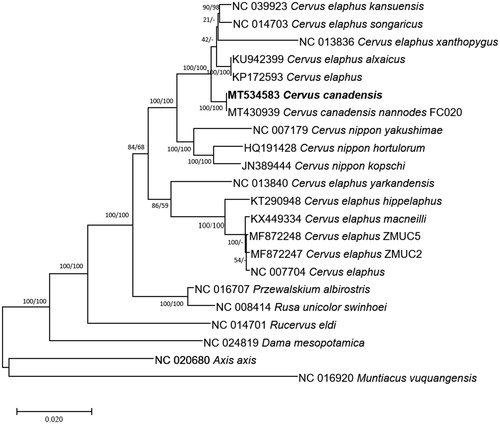
Twenty-two complete mitochondrial genomes of subfamily Cervinae and one outgroup species, Muntiacus vuquangensis, were used for constructing bootstrapped neighbor joining and maximum likelihood trees with MEGA X (Kumar et al. Citation2018) based on multiple alignment of complete mitochondrial genomes conducted by MAFFT 7.450 (Katoh and Standley Citation2013). Mitochondrial genomes of C. canadensis and C. canadensis nannodes are clustered together in one clade of both trees ().
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 30 complete mitochondrial genomes: Cervus canadensis (MT534583) used in this study, Cervus canadensis nannodes (MT430939), Cervus elaphus alxaicus (KU942399), Cervus elaphus (NC_007704 and KP172593), Cervus elaphus kansuensis (NC_039923), Cervus elaphus songaricus (NC_014703), Cervus elaphus yarkandensis (NC_013840), Cervus elaphus hippelaphus (KT290948), Cervus elaphus macneilli (KX449334), Cervus elaphus (MF872248 and MF872247), Cervus nippon yakushimae (NC_007179), Cervus nippon hortulorum (NC_013834), Cervus nippon hortulorum (HQ191428), Cervus nippon hortulorum (KR868807), Cervus nippon kopschi (JN389444), Dama mesopotamica (NC_024819), Przewalskium albirostris (NC_016707), Rucervus eldi (NC_014701), Rusa unicolor swinhoei (NC_008414), Axis axis (NC_020680), and Muntiacus vuquangensis (NC_016920) as an outgroup. Phylogenetic tree was drawn based on maximum likelihood tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitochondrial genome sequence can be accessed via accession number MT534583 in NCBI GenBank.
Additional information
Funding
References
- Benestad SL, Mitchell G, Simmons M, Ytrehus B, Vikøren T. 2016. First case of chronic wasting disease in Europe in a Norwegian free-ranging reindeer. Vet Res. 47(1):88.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim T-Y, Shon H-J, Joo Y-S, Mun U-K, Kang K-S, Lee Y-S. 2005. Additional cases of chronic wasting disease in imported deer in Korea. J Vet Med Sci. 67(8):753–759.
- Kim H-J, Hwang J-Y, Park K-J, Park H-C, Kang H-E, Park J, Sohn H-J. 2020. The first complete mitogenome of Cervus canadensis nannodes (Merriam, 1905). Mitochondrial DNA Part B. 5(3):2294–2296.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- McCullough DR, Fischer JK, Ballou JD. 1996. From bottleneck to metapopulation: recovery of the tule elk in California. In: Dale McCullough (ed.) Metapopulations and wildlife conservation; Washington, DC: Island Press. p. 375–403.
- Park J, Kim Y, Xi H. 2019. The complete chloroplast genome sequence of male individual of Korean endemic willow, Salix koriyanagi Kimura ex Goerz (Salicaceae). Mitochondrial DNA Part B. 4(1):1619–1621.
- Park J, Kim Y, Xi H, Heo K-I, Min J, Woo J, Lee D, Seo Y, Kim YH. 2020. The complete chloroplast genomes of two cold hardness coffee trees Coffea arabica L. (Rubiaceae). Mitochondrial DNA Part B. 5(2):1619–1621.
- Park J, Kim Y, Xi H, Kwon W, Kwon M. 2019. The complete chloroplast and mitochondrial genomes of Hyunsasi tree, Populus alba × Populus glandulosa (Salicaceae). Mitochondrial DNA Part B. 4(2):2521–2522.
- Seo BY, Jung JK, Ho Koh Y, Park J. 2019. The complete mitochondrial genome of Laodelphax striatellus (Fallén, 1826)(Hemiptera: Delphacidae) collected in a southern part of Korean peninsula. Mitochondrial DNA Part B. 4(2):2242–2243.
- Sohn H-J, Kim J-H, Choi K-S, Nah J-J, Joo Y-S, Jean Y-H, Ahn S-W, Kim O-K, Kim D-Y, Balachandran A. 2002. A case of chronic wasting disease in an elk imported to Korea from Canada. J Vet Med Sci. 64(9):855–858.
- Williams E. 2005. Chronic wasting disease. Vet Pathol. 42(5):530–549.
- Williams E, Young S. 1980. Chronic wasting disease of captive mule deer: a spongiform encephalopathy. J Wildl Dis. 16(1):89–98.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2.
