Abstract
The tea weevil, Myllocerinus aurolineatus (Voss), is a serious pest of tea plants. We have obtained and annotated the complete mitochondrial genome of M. aurolineatus (GenBank accession No. MH197100). The entire mt genome is 17,762 bp long with an A + T content of 75.45%. The mt genome of M. aurolineatus encodes all 37 genes that are typically found in animal mt genomes, consists of 13 protein-coding genes, 2 ribosomal RNA and 22 transfer RNA genes. The gene order is consistent with other weevil mt genomes in Entiminae, within a typical gene order of “RANSEF”. Phylogenetic analysis was performed using 13 protein-coding genes among 18 weevils showed that M. aurolineatus is closely related to another Entiminae species, Sympiezomias velatus.
The tea weevil, Myllocerinus aurolineatus (Voss), is one of the main leaf-feeding insects of Camellia sinensis. The adult weevils feeding on young, tender leaves, seriously deteriorating tea yield and quality (Sun et al. Citation2010). In this study, we obtained and described the complete mitochondrial genome of M. aurolineatus for the first time. Adult specimens were collected from a tea plantation at Yongchuan, Chongqing, China (N29°23′, E105° 54′), that the tea plantation is belong to Chongqing Academy of Agricultural Science. Samples have been deposited in the insect specimen room of Tea Research Institute of Chongqing Academy of Agricultural Science with an accession number of CQNKY-CO-01-01-01.
The complete mt genome of the tea weevil is a typical closed-circular and double stranded DNA molecule with 17,762 bp in length (GenBank accession MH197100). The overall nucleotide composition of the major strand of the mt genome as follows: A = 39.55% (7025), C = 14.53% (2580), G = 10.02% (1780) and T = 35.90% (6377), with a total A + T content of 75.45%, that is heavily biased toward A and T nucleotides. AT- and GC-skew of the whole J-strand of M. aurolineatus is 0.048 and −0.183, respectively. The mt genome of M. aurolineatus encodes all 37 genes usually found in animal mt genomes, including 13 protein-coding genes, 2 ribosomal RNAs and 22 transfer RNAs. In this mt genome, there are two long non-coding region like control region. One non-coding region locates between rrnS and trnI, is 1161 bp long with a high A + T content of 82.26%, and the other one locates between trnL and trnQ, is 2012 bp long with a low A + T content of 69.23%. The gene arrangement of the tea weevil is identical to other weevil mt genome in Entiminae, contains gene order “RANSEF” instead of the ancestral order “ARNSEF” (Tang et al. Citation2017). Twenty-three of all 37 gene are encoded on the majority strand (J-strand) and the others encoded by the minority strand (N-strand).
Twelve of the 13 PCGs start with ATN codons (ATG for atp6, cox3, cob and nad4L; ATT for cox2, nad2, nad3, nad5 and nad6; ATA for nad1 and nad4; ATC for atp8) and cox1 used AAT as start codon, same situation exists in the mt genome of Naupactus xanthographus (Song et al. Citation2010). Three PCGs (cox1, nad4 and nad5) have incomplete terminal codons consisting of single T nucleotide, and the other PCGs stop with TAA and TAG. The nucleotide length of tRNA genes is ranging from 63 bp (trnC, trnH and trnN) to 71 bp (trnK), and A + T content is ranging from 62.69% (trnS1) to 87.50% (trnC and trnD). Twenty-one tRNA genes have the conventional cloverleaf shaped secondary structure and trnS1 gene lacks the dihydrouridine (DHU) arm. The gene trnS1 almost lacks the DHU arm in all metazoan mt genomes (Cameron Citation2014). The two rRNA genes have been identified on the N-strand in the M. aurolineatus mt genome. The length of rrnL and rrnS genes was 1288 bp and 786 bp, and their A + T content was 80.51 and 79.13%, respectively.
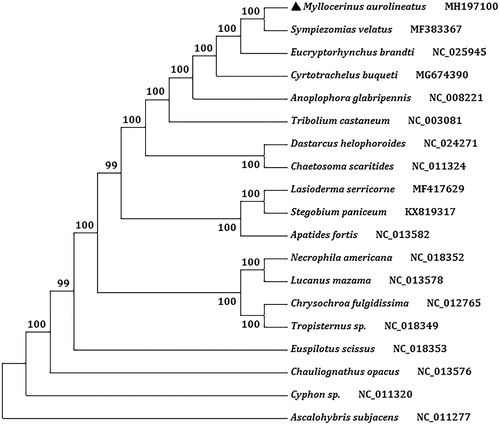
We analyzed amino acid sequence of 13 PCGs with maximum likelihood (ML) method to understand the phylogenetic relationship of M. aurolineatus with other weevils. The mt genome sequence of Ascalohybris subjacens (GenBank accession no. NC_011277) was used as an outgroup. The tea weevil and another Entiminae weevil, Sympiezomias velatus, are clustered into a branch of the phylogenetic tree with 100% bootstrap value ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MH197100.
Additional information
Funding
References
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Song HJ, Sheffield NC, Cameron SL, Miller KB, Whiting MF. 2010. When phylogenetic assumptions are violated: base compositional heterogeneity and among-site rate variation in beetle mitochondrial phylogenomics. Syst Entomol. 35(3):429–448.
- Sun XL, Wang GC, Cai XM, Jin S, Gao Y, Chen ZM. 2010. The tea weevil, Myllocerinus aurolineatus, is attracted to volatiles induced by conspecifics. J Chem Ecol. 36(4):388–395.
- Tang PA, Zhang L, Li XP, Li FF, Yuan ML. 2017. The complete mitochondrial genome of Sympiezomias velatus (Coleoptera: Curculionidae). Mitochondr DNA Part B Resour. 2(2):449–450.