Abstract
The complete mitochondrial genome (mitogenome) of the assassin bug, Sycanus croceovittatus, was sequenced and analyzed in the present study. This mitogenome spans 15,644 bp in size with a high A + T content (71.7%), containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a putative control region. All protein-coding genes are initiated by ATN codons expect ND1 use GTG as start codons and terminated with TAG or TAA, expect COX3 use a single T–– residue as the stop codon. All tRNAs have the typical clover-leaf like structures except for tRNASer(AGN). A phylogenetic analysis of S. croceovittatus and 33 other assassin bugs is also presented using 13 protein-coding genes and 2 rRNA genes. The result supports the monophyly of Harpactorinae and the sister relationship between S. croceovittatus and Agriosphodrus dohrni.
The assassin bugs Reduviidae is the second largest family in Heteroptera, including ∼7000 species ranked in almost 1000 genera and 25 subfamilies (Weirauch et al. Citation2014). In Reduviidae, the Harpactorinae is the largest subfamily distributed around the world mainly in tropical and subtropical regions. Here, we sequenced and described the complete mitochondrial genome (mitogenome) of Sycanus croceovittatus from subfamily Harpactorinae. Total genomic DNA was extracted from the samples collected from Xishuangbanna, Yunnan, China (21°56′N 101°15′E). Voucher specimen and the genomic DNA were stored at the Entomological Museum of China Agricultural University (No. HEM-019). The complete mitogenome was obtained by the next-generation sequencing method with Illumina Hiseq 2500 and the sequence has been submitted to the GenBank (Accession number: MT535605).
The complete mitogenome of S. croceovittatus is a single circular DNA molecule of 15,644 bp in length containing 13 protein-coding genes (PCGs), 22 transfer RNA (tRNAs), 2 ribosomal RNA (rRNAs), and a control region locating between srRNA and tRNAIle. The nucleotide composition of the whole mitogenome sequence is significantly biased towards A + T (71.7%) and exhibits positive AT-skew (0.10) and negative GC-skew (–0.16). Gene order is identical to the putative ancestral arrangement of insect (Boore Citation1999; Cameron Citation2014). In total, there are 52 bp overlapped nucleotides between neighboring genes in 13 locations, ranging from 1 to 14 bp in size.
The PCGs is 11,101 bp in length and nine PCGs are located on the majority strand (J strand) while the remaining genes are transcribed on the minority strand (N strand). All PCGs are initiated with ATN (two with ATA, two with ATC, two with ATT and six with ATG), except the ND1 uses the GTG as the start codon, and this unconventional start codon was also found in other assassin bugs (Li et al. Citation2012; Zhao, Jiang, et al. Citation2019). The typical stop codon TAA and TAG are assigned to 12 PCGs, whereas a single T residue is used by COX3 as stop codon which was repeatedly found in Reduviidae mitogenomes (Chen et al. Citation2018; Sun et al. Citation2019).
This mitogenome has the complete set of 22 tRNA genes, ranging from 61 to 71 bp. All tRNAs can be determined by the ARWEN (Laslett and Canbäck Citation2008) and folded into the typical clover-leaf structure except the tRNASer(AGN) with the dihydrouridine (DHU) arm formed a loop, which is commonly discovered in insect mitogenomes (Li et al. Citation2012; Song et al. Citation2019). Fourteen tRNAs are located on the J strand and the rest are encoded by the N strand. The length of lrRNA and srRNA is 1270 bp and 769 bp with A + T content of 75.4% and 72.7%, respectively. The largest noncoding region is the control region which is located between srRNA and tRNAIle with 932 bp in length. Except for the control region, there are 12 inter-genic regions ranging from 1 to 92 bp in size, of which the longest one is located between tRNASer(UCN) and ND1.
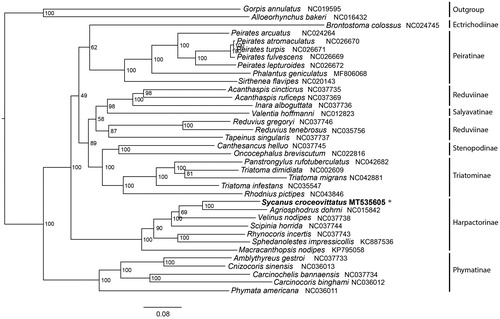
Maximum-likelihood (ML) tree was constructed by IQ-TREE 1.6.5 (Trifinopoulos et al. Citation2016) under the GTR + I + G model, using sequences of 13 protein-coding genes and 2 rRNA genes from 34 Reduviidae and 2 Nabidae species (). The phylogenetic relationships among Reduviidae is ((((Ectrichodiinae + Peiratinae) + ((Salyavatinae + Reduviinae) + (Stenopodinae + Triatominae))) + Harpactorinae) + Phymatinae). In the phylogenetic tree, each family forms a monophyletic cluster with high support except the subfamily Reduviinae. The subfamily Reduviinae was also suspected to be a paraphyly group in previous studies (Zhao, Chen, et al. Citation2019). The sister relationship between S. croceovittatus and Agriosphodrus dohrni is recovered with the high bootstrap support value.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study will be available in GenBank at https://www.ncbi.nlm.nih.gov/, Accession number MT535605.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Chen Z, Song F, Cai W. 2018. Complete mitochondrial genome of the ambush bug, Amblythyreus gestroi (Hemiptera: Reduviidae). Mitochondrial DNA Part B: Resources. 3(2):1044–1045.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Li H, Liu H, Song F, Shi A, Zhou X, Cai W. 2012. Comparative mitogenomic analysis of damsel bugs representing three tribes in the family Nabidae (Insecta: Hemiptera). PLOS One. 7(9):e45925.
- Song F, Li H, Liu GH, Wang W, James PD, Colwell D, Tran A, Gong SY, Cai WZ, Shao RF. 2019. Mitochondrial genome fragmentation unites the parasitic lice of eutherian mammals. Syst Biol. 68(3):430–440.
- Sun Z, Liu Y, Wilson JJ, Chen Z, Song F, Cai W, Li H. 2019. Mitochondrial genome of Phalantus geniculatus (Hemiptera: Reduviidae): trnT duplication and phylogenetic implications. Int J Biol Macromol. 129:110–115.
- Trifinopoulos J, Nguyen LT, Haeseler AV, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Weirauch C, Bérenger JM, Berniker L, Forero D, Forthman M, Frankenberg S, Freedman A, Gordon E, Hoey-Chamberlain R, Hwang WS, Marshall SA, et al. 2014. An illustrated identification key to assassin bug subfamilies and tribes (Hemiptera: Reduviidae). Can J Arthropod Identif. 26:1–115.
- Zhao Y, Chen Z, Song F, Li H, Cai W. 2019. The mitochondrial genome of the assassin bug Acanthaspis cincticrus (Hemiptera: Reduviidae). Mitochondrial DNA Part B. 4(1):474–475.
- Zhao Y, Jiang M, Wu Y, Song F, Cai W, Li H. 2019. Mitochondrial genomes of three kissing bugs (Reduviidae: Triatominae) and their phylogenetic implications. Int J Biol Macromol. 134:36–42.