Abstract
The complete mitochondrial genome (mitogenome) of Scythris sinensis Felder & Rogenhofer, 1775 (Lepidoptera: Scythrididae) was 15,216 bp with a typical set of genes (13 protein-coding genes [PCGs], 2 rRNA genes, and 22 tRNA genes) and one non-coding region, with an arrangement identical to that observed in most lepidopteran genomes. Twelve PCGs had the typical ATN start codon, whereas COI had the atypical CGA codon that is frequently found in the start region of the lepidopteran COI. The 271-bp long A + T-rich region was the shortest among sequenced Gelechioidea, which ranged from 290 – 375 bp. Phylogenetic analyses with concatenated sequences of the 13 PCGs, two RNA genes, and 22 tRNA genes using the Bayesian inference (BI) method, placed S. sinensis in the Scythrididae, as a sister to the family Stathmopodidae. The nodal support for this sister relationship was the highest at Bayesian posterior probabilities = 1.
Scythris sinensis Felder & Rogenhofer, 1775 (Lepidoptera: Scythrididae) belongs to Scythrididae. The species is found in eastern Asia including Korea, Europe, central Russia, southern Siberia, and eastern North America (Bengtsson Citation1997; Nupponen et al. Citation2000; Nupponen and Nupponen Citation2001; Landry et al. Citation2013; Bidzilya et al. Citation2017). The species occurs from May to July in Korea, has a pair of yellow patches on the front wings, and has bright yellow abdominal segments on its abdomen, which is broader in the female (Landry et al. Citation2013). The larvae of the species feed on Chenopodium album and Atriplex patula belonging to Chenopodiaceae and live in a loose spinning inside the young leaves at the top of the plant, concealed between the buds and leaves (Malkiewicz and Dobrzański Citation2011).
An adult male S. sinensis was collected from Gyeongju-city, Gyeongsangnam-do Province (35°50′42.0″N, 129°30′13.5″E), South Korea in 2012. This voucher specimen was deposited at the Chonnam National University, Gwangju, Korea, under the accession no. CNU6197. Using DNA extracted from the hind legs, three long overlapping fragments (LFs; COI-ND4, ND5-lrRNA, and lrRNA-COI) were amplified using three sets of primers that were previously published (Kim et al. Citation2012). Subsequently, these LFs were used as templates to amplify 26 short fragments (Kim et al. Citation2012).
Phylogenetic analysis was performed using the concatenated nucleotide sequences of 13 protein-coding genes (PCGs), two RNA genes, and 22 tRNA genes of 14 mitogenome sequences from Gelechioidea in Lepidoptera, including that of S. sinensis. The Bayesian inference (BI) method implemented in CIPRES Portal v. 3.1 (Miller et al. Citation2010) was used for phylogenetic analysis.
The complete 15,216-bp mitogenome of S. sinensis was composed of typical sets of genes (two rRNAs, 22 tRNAs, and 13 PCGs) and a major non-coding 271 bp A + T-rich region (GenBank accession no. MH230111). The gene arrangement of S. sinensis is identical to that of the ditrysian Lepidoptera that have the order trnM-trnI-trnQ (where underlining indicates a gene inversion) between the A + T-rich region and ND2 (Kim et al. Citation2010) instead of the ancestral trnI-trnQ-trnM order found in the majority of insects (Boore Citation1999). The A/T content of the whole mitogenome was 80.9%; however, it varied among the genes as follows: the A + T-rich region, 96.3%; srRNA, 85.1%; lrRNA, 84.2%; and PCGs, 79.5%. Twelve PCGs had the typical ATN start codon, whereas COI had the atypical CGA codon. Ten of the 13 PCGs had a complete stop codon (TAA); however, COI, COII, and ND5 had an incomplete stop codon, T.
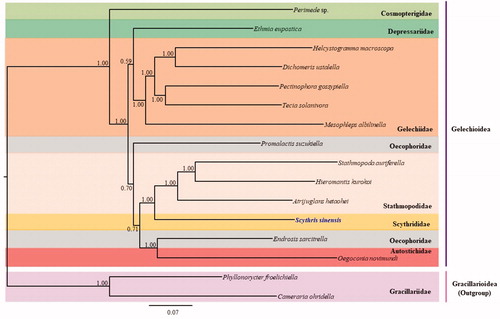
Phylogenetic analysis resulted in a sister relationship between Scythrididae, represented by the current S. sinensis and the Stathmopodidae, represented by Stathmopoda auriferella, Hieromantis kurokoi, and Atrijuglans hetaohei, with the highest nodal support (Bayesian posterior probabilities [BPP] = 1; ). This sister relationship also was supported by 19 nuclear gene-based analyses (Sohn et al. Citation2016). Among the three families in Gelechioidea represented by more than one species, Gelechiidae and Stathmopodidae formed strong monophyletic groups (BPP = 1), whereas Oecophoridae, represented by two species, formed a non-monophyletic group (). Currently, complete mitogenome sequences are only available from 14 species in Gelechioidea, including that of S. sinensis. Thus, more mitogenome sequences from such a diverse taxonomic group are required for further comprehensive phylogenetic inference.
Figure 1. Phylogenetic tree for Gelechioidea. The tree was constructed using nucleotide sequences of 13 protein-coding genes, two rRNA genes, and 22 tRNA genes via the Bayesian inference method. The numbers at each node indicate the Bayesian posterior probabilities. The scale bar indicates the number of substitutions per site. Two species belonging to the family Gracillariidae in Gracillarioidea were used as outgroups. GenBank accession numbers are as follows: Perimede sp., KJ508041 (Timmermans et al. Citation2014); Ethmia eupostica, KJ508047 (Timmermans et al. Citation2014); Helcystogramma macroscopa, KT354968 (Ma et al. Citation2016); Dichomeris ustalella, KU366706 (Park et al. Citation2016b); Pectinophora gossypiell, KM225795 (Zhao et al. Citation2016); Tecia solanivora, KT326187 (Ramírez-Ríos et al. Citation2016); Mesophleps albilinella, KU366707 (Park et al. Citation2016b); Promalactis suzukiella, KM875542 (Park et al. Citation2016c); Stathmopoda auriferella, KX138529 (Jeong et al. Citation2016); Hieromantis kurokoi, KU605775 (Park et al. Citation2016a); Atrijuglans hetaohei, KT581634 (Wang et al. Citation2016); Endrosis sarcitrella, KJ508037 (Timmermans et al. Citation2014); Oegoconia novimundi, KJ508036 (Timmermans et al. Citation2014); Phyllonorycter froelichiella, KJ508048 (Timmermans et al. Citation2014); and Cameraria ohridella, KJ508042 (Timmermans et al. Citation2014).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in Mendeley Data at http://dx.doi.org/10.17632/rdwz62m66w.1
Additional information
Funding
References
- Bengtsson BǺ. 1997. Scythrididae. In: Huemer P, Karsholt O, Lyneborg L, editors. Microlepidoptera of Europe. Vol. 2. Stenstrup: Apollo Books; p. 1–301.
- Bidzilya OV, Budashkin YI, Zhakov AV. 2017. Checklist of scythridid moths (Lepidoptera, Scythrididae) of Ukraine with description of two new species. Zootaxa. 4291(3):481–503.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Jeong SY, Park JS, Kim SS, Kim I. 2016. Complete mitochondrial genome of the gelechioid Stathmopoda auriferella (Lepidoptera: Stathmopodidae). Mitochondrial DNA B. 1(1):522–524.
- Kim JS, Park JS, Kim MJ, Kang PD, Kim SG, Jin BR, Han YS, Kim I. 2012. Complete nucleotide sequence and organization of the mitochondrial genome of eri-silkworm, Samia Cynthia ricini (Lepidoptera: Saturniidae). J Asia Pac Entomol. 15(1):162–173.
- Kim MJ, Wan X, Kim KG, Hwang JS, Kim I. 2010. Complete nucleotide sequence and organization of the mitogenome of endangered Eumenis autonoe (Lepidoptera: Nymphalidae). Afr J Biotechnol. 9:735–754.
- Landry J-F, Nazari V, deWaard JR, Mutanen M, Lopez-Vaamonde C, Huemer P, Hebert P. 2013. Shared but overlooked: 30 species of Holarctic Microlepidoptera revealed by DNA barcodes and morphology. Zootaxa. 3749:1–93.
- Ma L, Dong WW, Jiang GF, Wang X. 2016. The complete mitochondrial genome of Brachmia macroscopa (Lepidoptera: Gelechiidae) and its related phylogenetic analysis. J Insect Sci. 16(1):9.
- Malkiewicz A, Dobrzański X. 2011. Scythris sinensis (Felder & Rogenhofer, 1775)-the first record in Poland, and some new regional records of Scythrididae (Lepidoptera). Pol J Entomol. 80(3):517–521.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the 9th Gateway Computing Environments Workshop (GCE), New Orleans. p. 1–8.
- Nupponen K, Bengtsson BǺ, Kaitila J-P, Nupponen T, Junnilainen J, Olschwang V. 2000. The scythridid fauna of the southern Ural Mountains, with description of fourteen new species (Lepidoptera: Scythrididae). Entomol Fennica. 11(1):5–34.
- Nupponen K, Nupponen T. 2001. Notes on the scythridid fauna of the Altai Mountains, with description of four new species (Lepidoptera: Scythrididae). Entomol Fenn. 12:81–93.
- Park JS, Jeong SY, Kim SU, Kim I. 2016a. Complete mitochondrial genome of the gelechioid Hieromantis kurokoi (Lepidoptera: Stathmopodidae). Mitochondrial DNA B. 1(1):285–313.
- Park JS, Kim MJ, Jeong SY, Kim SS, Kim I. 2016b. Complete mitochondrial genomes of two gelechioids, Mesophleps albilinella and Dichomeris ustalella (Lepidoptera: Gelechiidae), with a description of gene rearrangement in Lepidoptera. Curr Genet. 62(4):809–826.
- Park JS, Kim SS, Kim KY, Kim I. 2016c. Complete mitochondrial genome of Suzuki’s Promolactis moth Promalactis suzukiella (Lepidoptera: Oecophoridae). Mitochondrial DNA Part A. 27:1–2094.
- Ramírez-Ríos V, Franco-Sierra ND, Alvarez JC, Saldamando-Benjumea CI, Villanueva-Mejía DF. 2016. Mitochondrial genome characterization of Tecia solanivora (Lepidoptera: Gelechiidae) and its phylogenetic relationship with other lepidopteran insects. Gene. 581(2):107–116.
- Sohn J-C, Regier JC, Mitter C, Adamski D, Landry J-F, Heikkilä M, Park K-T, Harrison T, Mitter K, Zwick A, Kawahara AY, et al. 2016. Phylogeny and feeding trait evolution of the mega-diverse Gelechioidea (Lepidoptera: Obtectomera): new insight from 19 nuclear genes. Syst Entomol. 41(1):112–132.
- Timmermans MJ, Lees DC, Simonsen TJ. 2014. Towards a mitogenomic phylogeny of Lepidoptera. Mol Phylogenet Evol. 79:169–178.
- Wang Q, Zhang Z, Tang G. 2016. The mitochondrial genome of Atrijuglans hetaohei Yang (Lepidoptera: Gelechioidea) and related phylogenetic analyses. Gene. 581(1):66–74.
- Zhao J, Sun Y, Xiao L, Tan Y, Dai H, Bai L. 2016. Complete mitochondrial genome of the pink bollworm Pectinophora gossypiella (Lepidoptera: Gelechiidae. Mitochondrial DNA A. 27(4):2833–1576.
