Abstract
Different species of Zingiberaceae are very similar in appearance, which brings some obstacles in the accurate identification of species. Here, we assembled and analyzed the complete chloroplast (cp) genome of Zingiber zerumbet. The cp genome of Z. zerumbet is 169,183 bp in length, which contains a pair of inverted repeated (IR) regions of 37,480 bp, a large single copy (LSC) region of 86,709 bp, and a small single copy (SSC) region of 7,515 bp. The genome contains a total of 138 genes, including 92 protein-coding genes, 38 transfer RNA genes, and eight ribosomal RNA genes. Phylogenetic analysis exhibited that the cp genome sequence could distinguish Z. zerumbet from other species. This study is beneficial to the germplasm identification and utilization of Z. zerumbet.
Zingiberaceae is a member of zingiberales, forming an isolated group among the monocotyledons. Of the 47 genera and 1400 species of Zingiberaceae, about 1000 species are distributed in tropical Asia (Abdul et al. Citation2008). Most Zingiberaceae plants have medicinal value, but there are some differences in their medicinal efficacy. Due to the similar appearance of Zingiberaceae, its evolution and systematic classification have been controversial. The development of DNA molecular markers helps to identify Zingiberaceae plants quickly and accurately.
Zingiber zerumbet is a vigorous Zingiberaceae plant, with leafy stems that can grow about 1.2 meters long (Kader et al. Citation2010). It is found in many tropical countries. The rhizomes of Z. zerumbet are used as food condiments and appetizers, while their root extracts are widely used in herbal medicine. Zerumbone, for example, is a natural chemotherapy drug derived from Z. zerumbet. It has anti-inflammatory, anti-hypersensitivity and anti-microbial activities, and can be used to treat many chronic diseases such as osteoarthritis, diabetic nephropathy, atherosclerosis, inflammation and cancer (Singh et al. Citation2019). The chloroplast genome plays an important role in species identification, However, the chloroplast genome of Z. zerumbet has not been reported. In this study, the complete chloroplast (cp) genome of Z. zerumbet was sequenced and identified using the Illumina sequencing technology. Total genomic DNA was extracted from fresh leaves of Z. zerumbet collected from Tunchang County, Hainan Province, China (19°19′31″N, 110°06′31″E). The specimens were deposited in the biotechnology department of Hainan academy of agricultural sciences (Specimen code HQJ002). A genomic shotgun library with an insertion size of 300 bp was constructed, the libraries were sequenced on Illumina HiSeq 4000 platform, then the clean data were de novo assembled using SPAdes v. 3.11.0 software (Bankevich et al. Citation2012). Finally, the assembled complete cp genome was annotated by Plann software (Huang and Cronk Citation2015) and submitted to GenBank under the accession number of MT239400.
The total length of complete cp genome of Z. zerumbet is 169,183 bp, with a total GC content of 35.8%. The complete cp genome has a typical quadripartite structure, including a large single copy (LSC) region of 86,709 bp, a small single copy (SSC) region of 7515 bp and two inverted repeat (IRS) regions of 37,480 bp. The complete cp genome contains 138 genes, including 92 protein-coding genes, 38 tRNA, and eight rRNA genes.
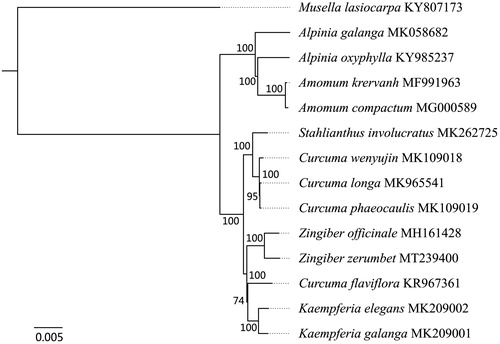
To determine the phylogenetic position of Z. zerumbet, the complete cp genome of Z. zerumbet was aligned with that of other 13 species in GenBank using HomBlocks software (Bi et al. Citation2018). Subsequently, the phylogenetic tree was constructed by the maximum likelihood method in RAxMLv8.2.9 software (Stamatakis Citation2014). The result indicated that Z. zerumbet is closely related to Zingiber officinale (). These information will provide useful genomic resources for the development of molecular markers and accurate identification of Z. zerumbet.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/MT239400.1/, reference number MT239400.
Additional information
Funding
References
- Abdul A, Al-Zubairi A, Tailan N, Wahab S, Zain Z, Ruslay S, Syam M. 2008. Anticancer activity of natural compound (Zerumbone) extracted from Zingiber zerumbet in human HeLa cervical cancer cells. Int J Pharmacol. 4(3):160–168.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Kader M, Habib M, Nikkon F, Yeasmin T, Rashid M, Rahman M, Gibbons S. 2010. Zederone from the rhizomes of Zingiber zerumbet and its anti-staphylococcal activity. Boletín Latinoamericano y Del Caribe de Plantas Medicinales y Aromáticas. 09:63–68.
- Singh YP, Girisa S, Banik K, Ghosh S, Swathi P, Deka M, Padmavathi G, Kotoky J, Sethi G, Fan L, et al. 2019. Potential application of zerumbone in the prevention and therapy of chronic human diseases. J Funct Foods. 53:248–258.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.