Abstract
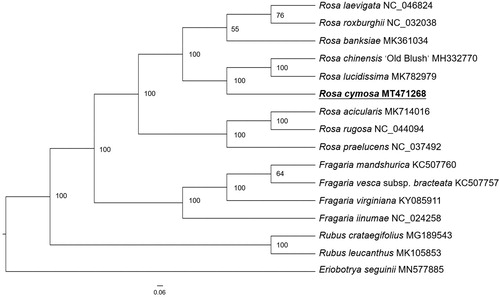
Rosa cymosa is a traditional medicinal and ornamental plant in China. Here, we report the complete chloroplast genome of R. cymosa. The chloroplast genome is 156,607 bp in length with 37.48% GC content, containing a small single-copy (SSC) region (18,763 bp), a large single-copy (LSC) region (85,722 bp), and a pair of inverted repeats (IRs: 26,061 bp each). A total of 139 genes were predicted, including 92 protein-coding genes, eight ribosomal RNA genes, and 39 tRNA genes. Phylogenetic analysis based on chloroplast genomes of 16 plant species shows that R. cymosa is closest to R. chiensis ‘Old Bush’ and R. lucidissima. These complete chloroplast genomes can be subsequently used for researches of Rosaceae.
Rosa cymosa Tratt. is an evergreen climbing or scandent shrub belongs to Rosaceae and is wildly distributed in the south region of China (Gu and Robertson Citation2003). Rosa cymosa is an important species with potential exploitation and utilization values. The roots are used in the treatment of diarrhea, rheumatoid arthritis, descensus uteri, and hemorrhage as a traditional Chinese herbal remedy (Takashi et al. Citation1993). The flowers are beautiful and fragrant with strong climbing ability of the branches, which can be used for vertical greening of gardens, buildings, etc. (Zappi and Taylor Citation1997; Liu et al. Citation2011). In this study, we assembled the complete chloroplast genome of R. cymosa and explored the phylogenetic relationship with other species in Rosacea family.
Fresh and young leaves of R. cymosa were collected from Nan’ao Island, Guangdong Province, China (N23°26’02.07”, E116°58’03.84”), and the voucher specimen was deposited in the Herbarium of South China Agricultural University (CANT) under the accession number 440523-190718-018. The total genomic DNA was extracted from fresh leaves using a modified CTAB method (Doyle and Doyle Citation1987). A genomic library consisting of an insert size of 300 bp was constructed using TruSeq DNA Sample Prep Kit (Illumina, USA), and sequencing was carried out on an Illumina HiSeq Nova platform (Guangzhou Jierui Biotech). 6 Gb raw data of 150 bp paired-end reads were obtained and further assembled using GetOrganelle (Jin et al. Citation2018). The cp genome annotation was accomplished using Geseq (Tillich et al. Citation2017) and then manually checked by comparison against the complete cp genome of R. rugosa (Genbank accession number: NC_044094). The complete chloroplast genome of R. cymosa was submitted to GenBank with the accession number MT471268.
The complete chloroplast genome of R. cymosa (MT471268) is 156,607 bp in size, containing a small single-copy (SSC) region (18,763 bp) and a large single-copy (LSC) region (85,722 bp), separated by a pair of inverted repeats (IRs: 26,061 bp each). The overall GC content of the cp genome is 37.48%. There are 139 genes reported, including 92 protein-coding genes, eight ribosomal RNA genes, and 39 tRNA genes.
For phylogenetic analysis, a maximum-likelihood (ML) tree was performed based on the complete chloroplast genome sequences of 16 species from Rosoideae subfamily of Rosaceae family, with Eriobotrya seguinii as outgroup. All of the sequences were downloaded from NCBI GenBank. The ML analysis was constructed by RAxML software (Stamatakis Citation2014) with 1000 bootstrap replicates. The ML tree () showed that R. cymosa was the sister clade to R. chiensis ‘Old Bush’ and R. lucidissima, the species from Fragaria and Rubus were clustered respectively as sister groups to Rosa. This finding could serve as a valuable genomic resource for genetic researches on Rosaceae in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number MT471268.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gu CZ, Robertson KR. 2003. Rosaceae. Flora China. 9:365–455.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv. :256479. https://doi.org/10.1101/256479
- Liu CC, Liu YG, Guo K, Li GQ, Zheng YR, Yu LF, Yang R. 2011. Comparative ecophysiological responses to drought of two shrub and four tree species from karst habitats of southwestern China. Trees. 25(3):537–549.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Takashi Y, Feng WS, Okuda T. 1993. Two polyphenol glycosides and tannins from Rosa cymosa. Phytochemistry. 32(4):1033–1036.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zappi D, Taylor N. 1997. Plate 309. Rosa cymosa ‘Rebecca Rushforth’. Curtis's Bot Mag. 14(1):13–15.