Abstract
Tapiscia sinensis, belong to Tapisciaceae, is endangered tree endemic to China. Here, we provide the complete plastid genomic data of T. sinensis with the aim of providing data for future conservation efforts research and revealing its phylogenetic position. The complete chloroplast sequence is 161,093 bp, including a large single-copy (LSC) region of 87,782 bp, a small single-copy (SSC) region of 18,517 bp, a pair of invert repeats (IR) regions of 27,387 bp. Plastid genome contains 131 genes, 85 protein-coding genes, 38 tRNA genes, and eight rRNA genes. Phylogenetic analysis base on 19 plastid genomes indicates that T. sinensis located Malvids branch, and is more closely related to the species of the order Sapindales than those of the order Malvales.
Keywords:
The Tapisciaceae are part of the Huerteales, which together with Geraniales, Myrtales, Crossosomatales, Picramniales, Sapinales, Malvales, and Brassicales, constitute the malvids (APG IV Citation2016). Tapiscia sinensis, belong to Tapisciaceae, is an ancient and endangered tree endemic to China, and mainly distributed in southwestern China at an elevation of 250–2200 m (Di and Yu Citation1989; Zhang Citation1989). Tapiscia sinensis is a good research object to infer the refugial location and colonization history of plant species in subtropical China because it is a Tertiary relict endemic to subtropical China (Zhang, Han, et al. Citation2018). Wood from T. sinensis is used in the production of furniture due to its light wood and beautiful texture (Wei et al. Citation2016). The leaves of T. sinensis are rich in flavonoids, which are widely used in medicinal materials (Xie Citation2006). Besides, T. sinensis are often used as ornamental trees because of their straight trunks and brightly colored flowers (Zhou et al. Citation2015). Although T. sinensis was listed as a state third class protected species in China in 1992, the natural population has been fragmented and threatened due to pollination difficulties, inability to bear fruit, weak natural regeneration, and human interference (Fu Citation1992; Ma Citation2013; Cai and Zhang Citation2017). It is imperative to provide theoretical data for effective conservation strategies for this important plant based on molecular data. Here, we report the complete plastid genome of T. sinensis based on Illumina pair-end sequencing technology. This information will be helpful to study the origin, evolution, and diversification of Tapisciaceae and other species.
The fresh leaves of T. sinensis were collected from Fuzhou Arboretum, Fuzhou City, Fujian province, China (119°17´32′´E, 26°08´56′´N), and used for DNA extraction (Qiu et al. Citation2020). The voucher specimen is kept at the Herbarium of College of Forestry, Fujian Agriculture and Forestry University (specimen code FAFU08018) (Yang et al. Citation2019). Total genomic DNA was extracted using the modified CTAB method, and the Covaris ultrasonic breaker was used to randomly interrupted 500 bp for library construction (Xiang et al. Citation2019). The constructed library was sequenced PE150 by Illumina Hiseq Xten platform, approximately 2 GB data was generated (Wang et al. Citation2019). Illumina data was filtered by script in the cluster (default parameter: -L 5, -p 0.5, -N 0.1). The plastid genome of T. sinensis was assembled by GetOrganelle pipe-line (https://github.com/Kinggerm/GetOrganelle) with complete plastid genome of Staphylea trifolia (GeneBank accession: MK488092) as reference (Chen et al. Citation2019). Then, we used Bandage to view and edit these sequences of assembly (Wick et al. Citation2015). Assembled plastid genome annotation was conducted base on comparison with S. trifolia by Geneious v 11.1.5 (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. Citation2012). The annotation result was drawn with the online tool OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013). The complete plastid genome sequence of T. sinensis has been submitted to GenBank with the accession number MT492024.
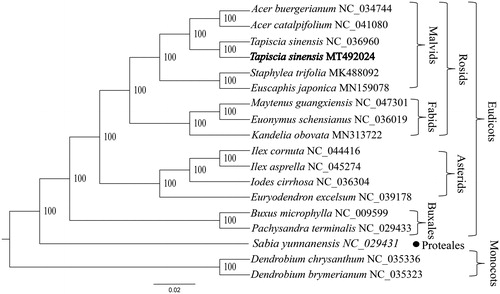
The complete plastid genome sequence of T. sinensis was 161,093 bp in length, with a large single-copy (LSC) region of 87,782 bp, a small single-copy (SSC) region of 18,517 bp, and a pair of inverted repeats (IR) regions of 27,387 bp. This result was basically consistent with the results reported by Zhang et al. (Zhang, Wang, et al. Citation2018). The complete plastid genome encodes 131 genes, containing 85 protein-coding genes, 38 transfer RNA (tRNA) genes, and eight ribosomal (rRNA) genes. The complete genome GC content was 37.2%. To reveal the phylogenetic position of T. sinensis, a phylogenetic tree was conducted based on complete plastid genomes of 18 species, including two Buxales species (Pachysandra terminalis and Buxus microphylla), one Proteales species (Sabia yunnanensis), three Fabids species (Kandelia obovata, Maytenus guangxiensis, and Euonymus schensianus), six Malvids species (two T. sinensis, Euscaphis japonica, S. trifolia, Acer catalpifolium, and A. buergerianum), four Asterids species (Euryodendron excelsum, Iodes cirrhosa, Ilex cornuta, and I. asprella), and two monocots species (Dendrobium chrysanthum and D. brymerianum) as outgroups. These genome data were downloaded from NCBI GenBank (Li et al. Citation2019). The sequences were aligned by MAFFT v7.307 with 1000 bootstrap replicates (Katoh and Standley Citation2013), and the phylogenetic tree constructed by RAxML (Stamatakis Citation2014). The phylogenetic tree showed that the Proteales and Buxales are located at the base of the eudicots branch, the remaining eudicots are divided into the Rosids branch and the Asterids branch. In addition, T. sinensis located Malvids branch, and is more closely related to the species of the order Sapindales than those of the order Malvales (). The completed plastid genome sequence of T. sinensis can help reveal its phylogenetic position, and provide data for future conservation efforts and biological research.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT492024.
Additional information
Funding
References
- APG IV. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Cai S, Zhang F. 2017. Study on cutting propagation technology of Tapiscia sinensis. Hubei Forestry Sci Technol. 46 (6):84–85.
- Chen DQ, Xiang S, Liu ZJ, Zou SQ. 2019. The complete chloroplast genome sequence of Kandelia obovata (Rhizophoraceae). Mitochondrial DNA B. 4(2):3494–3495.
- Di WZ, Yu ZY. 1989. The first countries to protect rare and endangered plants in Shaanxi province. Xi’an: Northwest University Press.
- Fu LK. 1992. China plant red data book: rare and endangered plants. Beijing: Science Press.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li J-W, Wang N, Yang Y, Wu X, Zou XX. 2019. The complete chloroplast genome sequence of Eranthis stellata (Ranunculaceae). Mitochondrial DNA B. 4(2):4025–4026.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Ma AM. 2013. Seedling raising and afforestation technology of Tapiscia sinensis. Anhui Forestry Sci Technol. 39(1):73–74.
- Qiu M-Y, Yang Y, Wang N, Wu X, Hu Y-L, Zou X-X. 2020. The re-sequencing of complete chloroplast genome of Cinnamomum camphora (Lauraceae) from Quanzhou, China. Mitochondrial DNA B. 5(1):520–521.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang N, Qiu M-Y, Yang Y, Li J-W, Zou X-X. 2019. Complete chloroplast genome sequence of Bougainvillea Spectabilis (Nyctaginaceae). Mitochondrial DNA B. 4(2):4010–4011.
- Wei HY, Liang HW, Zhang B, Li ZL. 2016. Establishment of embryogenic cell suspension system and plant regeneration of rare and endangered plant Tapiscia sinensis. Mol Plant Breed. 14(3):756–759.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Xiang S, Liu X-D, Sun W-H, Lan S-R, Liu Z-J, Zou S-Q. 2019. The complete chloroplast genome sequence of Euscaphis japonica (Staphyleaceae). Mitochondrial DNA B. 4(2):3484–3485.
- Xie CP. 2006. A review of research advances in rare and endangered plant Tapiscia sinensis. Subtrop Plant Sci. 35:71–74.
- Yang Y, Li J-W, Wang N, Zou X-X, Zou S-Q. 2019. The complete chloroplast genome sequence of Brasenia schreberi (Cabombaceae). Mitochondrial DNA B. 4(2):3842–3843.
- Zhang JX. 1989. Discovery of Tapiscia sinensis in Henan Province. Plant Journal. 1989(02):12–13.
- Zhang L, Han YZ, Liu ZQ, Meng YN, Liu YP, Kong DZ. 2018. Study on the growth rhythm of Tapiscia sinensis seedlings. Guangdong Agric Sci. 45 (02):47–51.
- Zhang PF, Wang ZW, Liu YL, Tian H, Yao XH, Zhang JL. 2018. Development and characterization of 11 polymorphic microsatellite markers in Tapiscia sinensis (Staphyleaceae). Application in plant sciences. Conserv Genet Resour. 10:765–768.
- Zhou XJ, Wang YY, Xu YN, Yan RS, Zhao P, Liu WZ. 2015. De novo characterization of flower bud transcriptomes and the development of EST-SSR markers for the endangered tree Tapiscia sinensis. Int J Mol Sci. 16(6):12855–12870.