Abstract
Punica granatum ‘Luqing1’ is an important d fruit tree with high quality and strong cold-resistance. The complete chloroplast genome (Accession: MN630638) was sequenced, assembled, and annotated. The size of chloroplast genome was 158,638 bp with large single-copy (LSC, 89,021 bp) regions, small single-copy (SSC, 18,685 bp) regions, and two inverted repeat regions (IRs, 25,466 bp each). A total of 130 genes were successfully annotated, including 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The phylogenetic relationships showed that P. granatum ‘Luqing1’ was closely related with different pomegranate cultivars.
Punica granatum L. is a deciduous fruit tree belongs to Lythraceae family and originates from Central Asia such as India and Iran (Feng et al. Citation2019). Pomegranate fruit is rich in soluble phenolic substances, amino acids, and ascorbic acid and is recognized worldwide for a pleasant taste and excellent health benefits (Karimi et al. Citation2017). It has strong adaptability and is cultivated widely in the world. ‘Luqing 1’ is an excellent cultivar of pomegranate, which has high quality, strong cold-resistance, and long storage time in China. The complete chloroplast (cp) genome is an efficient way to reveal the genetic characteristics and evolutionary relationship of valuable species (Cai et al. Citation2020). In this study, we characterized the complete chloroplast (cp) genome of P. granatum ‘Luqing1’ and performed phylogenetic analysis. It can provide theoretical foundation for the genetics conservation and evolution analysis of Punica genus.
The fresh leaves of P. granatum ‘Luqing1’ were collected from the Wanjishan field of Shandong Institute of Pomology (36.21°N, 117.08°E), Shandong Province, China. The voucher specimen accession was SDSL0083. Total genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Venlo, Netherlands). cpDNA sequencing was performed with an Illumina Hiseq 2500 platform by Nanjing Genepioneer Biotechnologies. The fastp program was used to filter the raw paired-end reads of cpDNA (Chen et al. Citation2018). The de novo assembly was performed by GetOrganelle (Jin et al. Citation2018) and annotated with DOGMA v1.2 program (Wyman et al. Citation2004). The OGDRAW was used to generate the circular genome map of the genome (Lohse et al. Citation2013). The complete cp genome of P. granatum ‘Luqing1’ was deposited in the GenBank (Accession: MN630638).
The complete cp genome of P. granatum ‘Luqing1’ was 158,638 bp in length with overall GC content of 36.92%. The cp genome was made up of LSC (Large single-copy region) with 89,021 bp, SSC (Small single-copy region) with 18,685 bp, and two IRs (Inverted repeat regions) with 25,466 bp. The whole complete cp genome encoded 130 unique genes, which contained 85 protein-coding genes, 37 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. The tRNA genes were distributed throughout the whole genome with 22 in the LSC, one in the SSC, and 14 in the IR regions, whereas rRNAs were only situated in the IR regions. There were 17 duplicated genes in IRs, including six protein-coding genes (ndhB, rpl2, rpl23, rps12, rps7, and ycf2), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and four rRNA (rrn16, rrn23, rrn4.5, and rrn5) genes. Among the protein-coding genes, three genes (clpP, rps12, and ycf3) contained two introns, and nine genes (atpF, ndhA, ndhB, petB, petD, rp116, rpoC1, and rps16) possessed a single intron, and six tRNA genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) also had one intron. There were 233 simple sequence repeats (SSRs) in total, which had 155 (66.50%) in LSC, 40 (17.20%) in SSC, and 38 (16.30%) in IR.
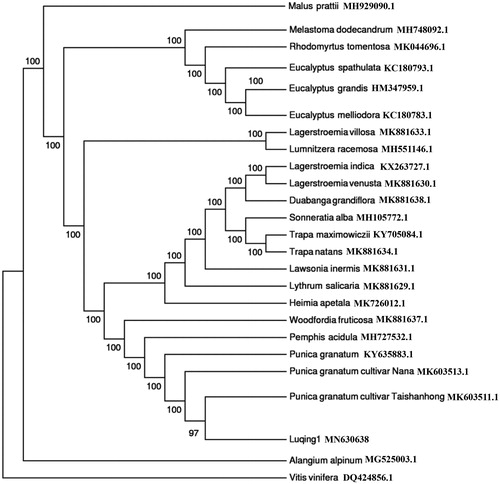
To ascertain the phylogenetic position of P. granatum ‘Luqing1’, 23 complete cp genomes within Lythraceae, Myrtaceae, and Melastomataceae family were selected. Malus prattii (GenBank Accession No. MH929090.1) and Vitis vinifera (GenBank Accession No. DQ424856.1) were as outgroup. The maximum-likelihood (ML) phylogenetic tree was constructed by the IQ-TREE with the best-fit model identified using ModelFinder (Kalyaanamoorthy et al. Citation2017). The result showed that P. granatum ‘Luqing1’ is closely related to the different pomegranate cultivars, such as ‘Taishanhong’ and ‘Nana’, and they had a close relationship with Pemphis acidula (). It will be valuable for the genetic study in Lythraceae family.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the NCBI at https://www.ncbi.nlm.nih.gov/, reference number: MN630638.
Additional information
Funding
References
- Cai Z, Wang H, Wang G. 2020. Complete chloroplast genome sequence of Pinellia ternata (Thunb.) Breit, a medicinal plants to China. Mitochondrial DNA B. 5(3):2107–2108.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Feng L, Yin Y, Yang X, Tang H, Jiao Q. 2019. Dynamic variations in punicalagin and related metabolic substances in pomegranate fruit and leaves during development periods. Hort J. 88(4):444–454.
- Jin JJ, Yu WB, Jun B, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 4:256479.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Karimi M, Sadeghi R, Kokini J. 2017. Pomegranate as a promising opportunity in medicine and nanotechnology. Trends Food Sci Tech. 69:59–73.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.