Abstract
The Hong Kong Dwarf Snake Calamaria septentrionalis Boulenger, 1890 is widely distributed in south of China and North of Vietnam. The complete mitochondrial genome (mitogenome) sequence of C. septentrionalis was determined by shotgun sequencing. The mitogenome of C. septentrionalis is 17,053 bp in length and contains two ribosome RNA genes, 13 protein-coding genes, 22 tRNA genes, and two noncoding regions. Most genes of C. septentrionalis were distributed on the H-strand, except for the ND6 subunit gene and eight tRNA genes which were encoded on the L-strand. The phylogenetic tree of C. septentrionalis and 15 other related species was built. The DNA data presented here will be useful to study the evolutionary relationships and genetic diversity of C. septentrionalis.
The genus Calamaria Boie, 1827 is one of the most diverse groups of Asian snakes with 64 known species (Grismer et al. Citation2006; Yang and Zheng Citation2018; Uetz et al. Citation2020). None complete mitochondrial genome was reported about this genus. Calamaria septentrionalis is widely distributed in south of China and North of Vietnam (Zhao et al., Citation1998; Zhao Citation2006). In this article, we determined and described the mitogenome of C. septentrionalis in order to obtain basic mitochondrial genetic information about this species.
The specimen of C. septentrionalis was collected from Tianzhushan Town (30°40′44″N, 116°28′32″E, 40 m a.s.l.) Qianshan County, Anhui Province, China. The sampling site is located in the southeast of Dabie Mountains. Muscle tissues were removed and immediately preserved in 75% ethanol. Total genomic DNA was extracted from muscle using a Qiagin DNEasy blood and tissue extraction kit (Qiagen Inc., Valencia, CA). This specimen and total genomic DNA were preserved and deposited in the Museum of Huangshan University (Voucher number: HS2020002). The complete mitogenome of C. septentrionalis (Genbank accession number MT478106) was sequenced to be 17,053 bp which consisted of two ribosome RNA genes, 13 typical vertebrate protein-coding genes, 22 transfer RNA (tRNA) genes, and two noncoding regions, which is similar to the typical mtDNA of other vertebrates (Boore Citation1999; Sorenson et al. Citation1999).
The annotation was predicted by the MITOS (Bernt et al. Citation2013), and the locations of protein-coding genes were corrected by comparing with the homologous genes of other closely related species. Most of the C. septentrionalis mitochondrial genes are encoded on the H-strand except for the ND6 gene and eight tRNA genes, which are encoded on the L-strand. The 12S rRNA (917 bp) and 16S rRNA (1427 bp), are located between the tRNA-Phe and protein-coding gene ND1 and separated by the tRNA-Val gene. Two noncoding control regions (D-loop) of the C. septentrionalis mitogenome in size are 668 bp and 1021 bp, respectively. The overall base composition of the entire genome was as follows: A (34.4%), T (25.4%), C (27.7%), and G (12.5%), which the percentage of G + C (40.2%) reflected a typical sequence feature of the vertebrate mitogenome.
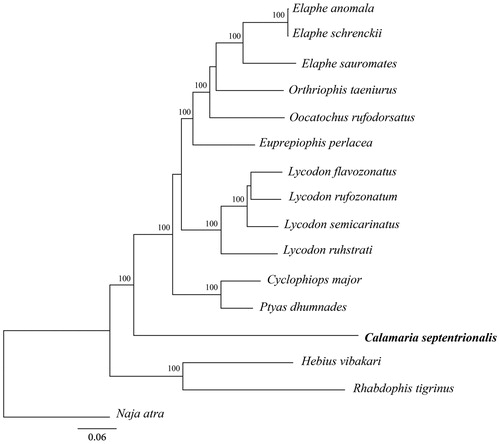
In order to validate the newly determined sequence, whole mitochondrial genome sequence of the S. incognitus determined in this study and together with other 15 related species from GeneBank to perform phylogenetic analysis. These species were as follows: Elaphe anomala, E. schrenckii, E. sauromates, Orthriophis taeniurus, Oocatochus rufodorsatus, Euprepiophis perlacea, Lycodon flavozonatus, L. rufozonatum, L. ruhstrati, L. semicarinatus, Cyclophiops major, Ptyas dhumnades, C. septentrionalis, Hebius vibakari, Rhabdophis tigrinus, Naja atra. A maximum-likelihood (ML) tree was constructed based on the dataset by online tool RAxML (Kozlov et al. Citation2019). The phylogenetic analysis result shows that C. septentrionalis resides within the clade representing the family Colubridae (). It indicated that the new determined mitogenome sequences could meet the demands and explain some evolution issues.
Figure 1. A maximum-likelihood (ML) tree of the Calamaria septentrionalis in this study and other 15 related species was constructed based on the dataset of the whole mitochondrial genome by online tool RAxML. The numbers above the branch meant bootstrap value. Bold fonts highlighted the study species and corresponding phylogenetic classification. The analyzed species and corresponding NCBI accession number as follows: Elaphe anomala (KP900218), E. schrenckii (KP888955), E. sauromates (MK070315), Orthriophis taeniurus (KC990021), Oocatochus rufodorsatus (KC990020), Euprepiophis perlacea (KF750656), Lycodon flavozonatus (KR911720), L. rufozonatum (KF148622), L. ruhstrati (KJ179951), L. semicarinatus (AB008539), Cyclophiops major (KF148620), Ptyas dhumnades (KF148621), Calamaria septentrionalis (MT478106), Hebius vibakari (KP684155), Rhabdophis tigrinus (KU641019), Naja atra (EU921898).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT478106.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Grismer LL, Grismer JL, Mcguire JA. 2006. A new species of reed snake of the genus Calamaria H. Boie, 1827, from Pulau Tioman, Pahang, West Malaysia. Hamadryad. 1305(1):1–6.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21), 4453-4455.
- Sorenson MD, Ast JC, Dimcheff DE, Yuri T, Mindell DP. 1999. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol Phylogenet Evol. 12(2):105–114.
- Uetz P, Freed P, Hošek J. 2020. The reptile database. [accessed 2018 Sep 22]. http://reptile-database.org
- Yang JH, Zheng X. 2018. A new species of the genus Calamaria (Squamata: Colubridae) from Yunnan Province, China. Copeia. 106(3):485–491.
- Zhao EM. 2006. Snakes of China. Hefei (China): Anhui Sciences and Technology Publishing House; p. 184. (in Chinese).
- Zhao EM, Huang MH, Zong Y, Jiang YM, Huang QY, Zhao H, Ma JF, Zheng J, Huang ZJ, Wei G, et al. 1998. Fauna Sinica. (Reptilia vol. 3 Squamata: Serpentes). Beijing: Science Press (in Chinese).
