Abstract
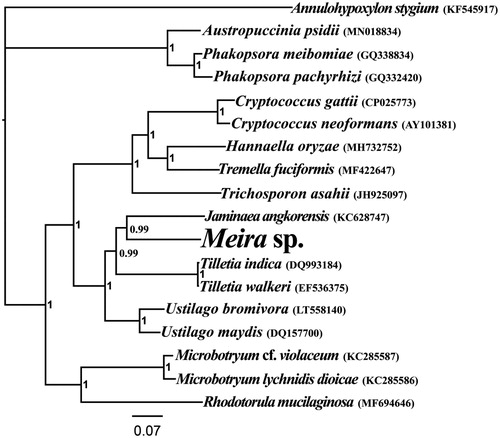
In this study, the complete mitochondrial genome of Meira sp. was sequenced and assembled. The complete mitochondrial genome of Meira sp. has 15 protein-coding (PCG) genes, 2 ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes. The mitochondrial genome of Meira sp. has a total size of 23,353 bp, with the base composition as follows: A (30.62%), T (32.82%), G (17.84%) and C (18.73%). Phylogenetic analysis indicated that the mitogenome of Meira sp. exhibited a close relationship with the mitogenome of Jaminaea angkorensis.
The genus Meira was first proposed to accommodate two anamorphic yeastlike fungi, namely Meira geulakonigii and Meira argovae (Boekhout et al. Citation2003). Meira species are usually distributed in plant tissues or on the leaf surface of plants (Yasuda et al. Citation2006; Limtong et al. Citation2017). So far, several molecular markers have been used in phylogenetic studies of Meira species, including nuclear rRNA genes, the D1/D2 region of the LSU rRNA gene, the SSU rRNA gene, and so on (Rush and Aime Citation2013; Wang et al. Citation2015). However, no complete mitochondrial genome from the genus Meira has been published. The mitochondrial genome of Meira sp. will promote the understanding of the taxonomy, phylogeny and evolution of the anamorphic yeast-like fungi.
The specimen (Meira sp.) was isolated from a plant tissue in Yanbian, Jilin, China (129.32 E; 43.11 N). The specimen was stored in Culture Collection Center of Chengdu University (No. Msp003). The total genomic DNA of Meira sp. was extracted using a Fungal DNA Kit D3390-00 (Omega Bio-Tek, Norcross, GA, USA) according to the instructions. We purified the extracted total DNA using a Gel Extraction Kit (Omega Bio-Tek, Norcross, GA, USA). Purified DNA was stored in Chengdu University (No. DNA_ Msp003). We constructed sequencing libraries with purified genomic DNA using the NEBNext® Ultra™ II DNA Library Prep Kit (NEB, Beijing, China). Whole genomic sequencing (WGS) was conducted by the Illumina HiSeq 2500 Platform (Illumina, SanDiego, CA). We The de novo assembled the complete mitogenome of Meira sp. using SPAdes 3.9.0 (Bankevich et al. Citation2012). The obtained complete mitogenome of Meira sp. was annotated according to the methods described by Li et al. (Li, Chen, et al. Citation2018; Li, Liao, et al. Citation2018; Li, Ren, et al. Citation2019; Li, Wang et al. Citation2018).
The complete mitogenome of Meira sp. is 23,353 bp in length, with the base composition as follows: A (30.62%), T (32.82%), G (17.84%) and C (18.73%). The complete mitogenome contains 15 protein-coding genes, 2 ribosomal RNA genes (rns and rnl), and 22 transfer RNA (tRNA) genes. To investigate the phylogenetic status of Meira sp., we constructed a phylogenetic tree for 18 species. We constructed the phylogenetic tree using Bayesian analysis (BI) method based on the combined 14 core protein-coding genes according to methods described by Li et al (Li, Wang, Jin, Chen, Xiong, Li, Liu, et al. Citation2019; Li, Wang, Jin, Chen, Xiong, Li, Zhao, et al. Citation2019; Li et al. Citation2020). As shown in the phylogenetic tree (), the mitogenome of Meira sp. exhibited a close relationship with the mitogenome of Jaminaea angkorensis (Hegedusova et al. Citation2014).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
This mitogenome of Meira sp. was submitted to GenBank under the accession of MT556837 (https://www.ncbi.nlm.nih.gov/nuccore/MT556837).
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Boekhout T, Theelen B, Houbraken J, Robert V, Scorzetti G, Gafni A, Gerson U, Sztejnberg A. 2003. Novel anamorphic mite-associated fungi belonging to the Ustilaginomycetes: Meira geulakonigii gen. nov., sp. nov., Meira argovae sp. nov. and Acaromyces ingoldii gen. nov., sp. nov. Int J Syst Evol Microbiol. 53(Pt 5):1655–1664.
- Hegedusova E, Brejova B, Tomaska L, Sipiczki M, Nosek J. 2014. Mitochondrial genome of the basidiomycetous yeast Jaminaea angkorensis. Curr Genet. 60(1):49–59.
- Li Q, Chen C, Xiong C, Jin X, Chen Z, Huang W. 2018. Comparative mitogenomics reveals large-scale gene rearrangements in the mitochondrial genome of two Pleurotus species. Appl Microbiol Biotechnol. 102(14):6143–6153.
- Li Q, Liao M, Yang M, Xiong C, Jin X, Chen Z, Huang W. 2018. Characterization of the mitochondrial genomes of three species in the ectomycorrhizal genus Cantharellus and phylogeny of Agaricomycetes. Int J Biol Macromol. 118:756–769.
- Li Q, Ren Y, Shi X, Peng L, Zhao J, Song Y, Zhao G. 2019. Comparative mitochondrial genome analysis of two ectomycorrhizal fungi (Rhizopogon) reveals dynamic changes of intron and phylogenetic relationships of the subphylum Agaricomycotina. Int. J. Mol. Sci. 20:5167–5182.
- Li Q, Wang Q, Chen C, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W. 2018. Characterization and comparative mitogenomic analysis of six newly sequenced mitochondrial genomes from ectomycorrhizal fungi (Russula) and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol. 119:792–802.
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Liu Q, Huang W. 2019. Characterization and comparative analysis of six complete mitochondrial genomes from ectomycorrhizal fungi of the Lactarius genus and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol. 121:249–260.
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W. 2019. Characterization and comparison of the mitochondrial genomes from two Lyophyllum fungal species and insights into phylogeny of Agaricomycetes. Int J Biol Macromol. 121:364–372.
- Li Q, Yang L, Xiang D, Wan Y, Wu Q, Huang W, Zhao G. 2020. The complete mitochondrial genomes of two model ectomycorrhizal fungi (Laccaria): features, intron dynamics and phylogenetic implications. Int J Biol Macromol. 145:974–984.
- Limtong S, Polburee P, Chamnanpa T, Khunnamwong P, Limtong P. 2017. Meira siamensis sp. nov., a novel anamorphic ustilaginomycetous yeast species isolated from the vetiver grass phylloplane. Int J Syst Evol Microbiol. 67(7):2418–2422.
- Rush TA, Aime MC. 2013. The genus Meira: phylogenetic placement and description of a new species. Antonie Van Leeuwenhoek. 103(5):1097–1106.
- Wang QM, Begerow D, Groenewald M, Liu XZ, Theelen B, Bai FY, Boekhout T. 2015. Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Stud Mycol. 81:55–83.
- Yasuda F, Izawa H, Yamagishi D, Akamatsu H, Kodama M, Otani H. 2006. Meira nashicola sp. nov., a novel basidiomycetous, anamorphic yeast-like fungus isolated from Japanese pear fruit with reddish stain. Mycoscience. 47(1):36–40.