Abstract
Dalbergia odorifera belongs to the family Fabaceae the genus Dalbergia that has high economic and medicinal value in China. The complete chloroplast genome of D. odorifera was 156,062 bp in length that had circular structure. It has the typical quadripartite structure of angiosperms, which includes 85,805 bp large single-copy region (LSC), 18,855 bp small single-copy region (SSC) of and a pair of 25,701 bp inverted repeat (IR) regions. The overall nucleotide composition of chloroplast genome sequence is 31.9% A, 32.0% T, 17.9% C, 18.2% G, and the total GC content of 36.1%. The chloroplast genome of D. odorifera annotated 129 genes, including 84 protein-coding genes, 37 transfer RNA genes, and eight ribosomal RNA genes. The phylogenetic analysis revealed that D. odorifera was close relationship to Dalbergia hainanensis in the family Fabaceae the genus Dalbergia using the maximum-likelihood (ML) method in this study.
Dalbergia odorifera belongs to the family Fabaceae the genus Dalbergia that has high economic and medicinal value in China. It is also a rare and endangered tree species endemic to Hainan Island, which produces the most expensive and rarest wood in China (Qin et al. Citation2020). The heartwood of D. odorifera named ‘Jiang-Xiang’ in Chinese as one kind of the traditional Chinese medicine (TCM) possess diverse pharmacological activities including antioxidant, anti-microbial and anti-inflammatory in China (Lee et al. Citation2014). In this study, the complete chloroplast genome of D. odorifera was assembled and completed that will provide more Traditional Chinese Medicine (TCM) genome information for future research and utilization.
Fresh specimens of D. odorifera were collected from the herb market near Beijing University of Chinese Medicine that located at Beijing (116.17E, 39.71 N) and preserved in liquid nitrogen. The total genomic DNA of D. odorifera was stored in Beijing University of Chinese Medicine (No. BJUCM-02). The complete chloroplast genome DNA was extracted from the fresh specimens and sequenced. The quality controlled and removed to the collected raw sequences using FastQC (Andrews Citation2015). The chloroplast genome of D. odorifera was assembled using MitoZ (Meng et al. Citation2019). Annotation was performed using Geneious (Kearse et al. Citation2012). All the coding and other genes in chloroplast genome of D. odorifera were predicted using CPGAVAS (Liu et al. Citation2012). Finally, the circular genome map was generated using DOGMA (Wyman et al. Citation2004).
The complete chloroplast genome of D. odorifera was 156,062 bp in length that had circular structure. It has the typical quadripartite structure of angiosperms, which includes 85,805 bp large single-copy region (LSC), 18,855 bp small single-copy region (SSC) of and a pair of 25,701 bp inverted repeat (IR) regions. The overall nucleotide composition of chloroplast genome sequence is 31.9% A, 32.0% T, 17.9% C, 18.2% G, and the total GC content of 36.1%. The chloroplast genome of D. odorifera annotated 129 genes, including 84 protein-coding genes, 37 transfer RNA genes, and eight ribosomal RNA genes. Among these, seven genes contain a single intron while two genes possess two introns. Seven protein-coding genes, seven tRNAs genes, and four rRNAs genes were duplicated in both IR regions. The chloroplast genome of D. odorifera had been deposited into the GenBank with the accession number NK9437781.
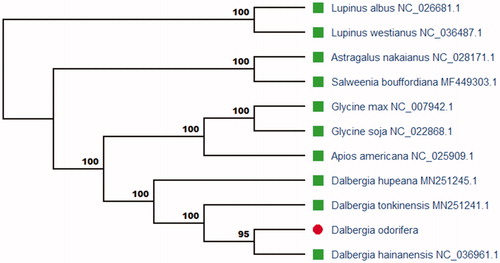
To explore the evolution status and phylogenetic relationship of D. odorifera, we based on the complete chloroplast genomes of 11 plant species complete chloroplast genome sequences to study. 11 plant species complete chloroplast genomes sequences were aligned using MAFFT (Katoh and Standley Citation2013) and conducted by maximum-likelihood (ML) method using MEGA X (Kumar et al. Citation2018). ML analysis method based on the Tamura–Nei model and 2000 bootstraps values was used for all the nodes. The ML phylogenetic tree () was drawn and edited using MEGA X. The phylogenetic analysis revealed that D. odorifera was close relationship to Dalbergia hainanensis in the family Fabaceae the genus Dalbergia. This study will provide more Traditional Chinese Medicine (TCM) genome information for future research and utilization.
Data openly available in a public repository that issues datasets with DOIs
The data that support the findings of this study are openly available in D. odorifera at http://doi.org/10.1080/23802359.2020.1781579
Disclosure statement
No potential conflict of interest was reported by all the authors. This study does not contain any studies with human participants or animals performed by any of the authors.
Data availability statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.,
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lee DS, Kim KS, Ko W, Li B, Keo S, Jeong GS, Oh H, Kim YC. 2014. The neoflavonoid latifolin isolated from meoh extract of Dalbergia odorifera attenuates inflammatory responses by inhibiting nf-κb activation via nrf2-mediated heme oxygenase-1 expression. Phytother Res. 28(8):1216–1223.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715–2164.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Qin M, Zhu C-J, Yang J-B, Vatanparast M, Schley R, Lai Q, Zhang D-Y, Tu T-Y, Klitgård BB, Li S-J, et al. 2020. Comparative analysis of complete plastid genome reveals powerful barcode regions for identifying wood of Dalbergia odorifera and D. tonkinensis (Leguminosae). J Syst Evol. doi:10.1111/jse.12598.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.