Abstract
The complete mitochondrial genome of Discogobio macrophysallidos was first determined and analyzed in this work. Its mitochondrial genome is 16,593 bp in length, consisting of 13 protein-coding genes, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and a non-coding control region, and its gene order was consistent with other fishes. Phylogenetic analysis showed that D. macrophysallidos were clustered with other three species of Discogobio. The complete mitogenome of D. macrophysallidos provides new molecular data for the further phylogenetic study of the genus of Discogobio.
Discogobio macrophysallidos is a small fish that belongs to subfamily Labeoninae (Teleostei: Cyprinidae), with special adaptability to rush water environment, and is a Yunnan-Guizhou plateau endemic species in China. In the past, the research on the genus Discogobio mostly focused on the comparison of external morphological features, such as the difference of disk and horseshoe-shaped skin fold, but did not involve the origin and phylogenetic research of this genus (Zhou et al. Citation2005; Zheng and Zhou Citation2008). Some studies have found that the interspecific morphology of this genus are similar, resulting in that there are heterologous with same name of some species (Zhao et al. Citation2017). In this study, we determined the complete mtDNA sequence of D. macrophysallidos for the first time, and assessed phylogenetic position with Labeoninae.
The sample was captured from Xingyun Lake (24°21′N, 102°46′E), Yunnan, China. Voucher specimens (No. 578) were deposited at the biological museum of Southwest Forestry University. Paired-end reads were sequenced using Illumina MiSeq platform. A total of 2,362,717 paired-end reads were obtained and assembled de novo. The tRNAs were confirmed using tRNAscan-SE (Schattner et al. Citation2005). The complete mitochondrial genome of D. macrophysallidos is 16,593 bp (GeneBank Accession No. MT536775), consisting of 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, 13 protein-coding genes, and a control region and the overall GC content was 41.94%, showing 96.51% identities to the Discogobio longibarbatus (GenBank: KY465995) (Zheng and Yang Citation2017). The order and direction of these genes were identical to other Labeoninae fishes (Xue et al. Citation2016; Zheng and Yang Citation2017; Tan et al. Citation2019; Wang et al. Citation2019).
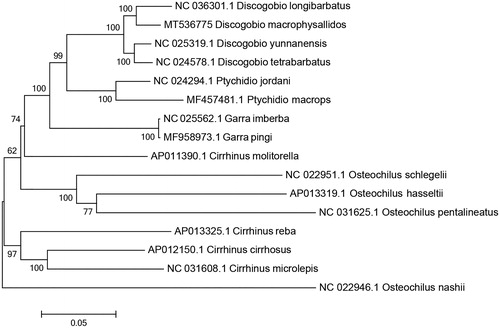
We used MEGA7 software (Sudhir et al. Citation2016) to construct a maximum-likelihood tree (with 1000 bootstrap replicates) based on complete mitogenomes of 16 species from Labeoninae subfamily. The phylogenetic tree showed that four species from Discogobio genus were clustered a clade with high bootstrap value supported (). The complete mitogenome of the D. macrophysallidos in this study provides important data in evolutionary analysis of Labeoninae phylogeny.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MT536775, reference numberMT536775.
Additional information
Funding
References
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689.
- Sudhir K, Glen S, Koichiro T. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Bio E. 33(7):1870–1874.
- Tan H, Zhang M, Cheng G, Chen X. 2019. The complete mitochondrial genome of Discogobio laticeps (Cyprinidae: Labeoninae). Mitochondrial DNA B Resour. 4(1):1981–1982.
- Wang W, Li S, Liu T, Wang X, Zeng S, Min W, Zhou L. 2019. Characterization of the complete mitochondrial genome of an endangere fish Semilabeo obscures (Cyrinidae, Lbeoninae, Semilabeo). Conserv Genet Resour. 11(2):147–150.
- Xue Y, Chen H-J, Li Y, Tang M, Chen H-J, Ye Q, Li Y. 2016. Mitochondrial genome of the sucking disc gudgeon, Discogobio yunnanensis (Teleostei, Cypriniformes). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1451–1452.
- Zhao JR, Zhao YY, Ge HL, Su SQ, Wang ZJ. 2017. Anatomical, histological observations of digestive system and digestive enzyme activity in Discogobio ynnanensis. Acta Hydrobiol Sin. 41(4):853–859.
- Zheng L, Yang J. 2017. Characterization of the complete mitochondrial genome of Discogobio longibarbatus (Cypriniformes, Cyprinidae), an endangered and endemic species from China. Conservation Genet Resour. 9(3):447–449.
- Zheng LP, Zhou W. 2008. Revision of the cyprinid genus Discogobio Lin, 1931 (Pisces: Teleostei) from the upper Red River basin in Wenshan Prefecture, Yunnan, China, with descriptions of three new species. Environ Biol Fish. 81(3):255–266.
- Zhou W, Pan XF, Kottelat M. 2005. Species of Garra and Discogobio (Teleostei: Cyprinidae) in Yuanjiang (Upper Red River) drainage of Yunnan province, China with description of a new species. Zool Stu. 44(4):445–453.