Abstract
The complete chloroplast genome of Russian sage Salvia yangii B. T. Drew was assembled in this study. The genome is 151,473 bp in length and contained 129 encoded genes in total, including 84 protein-coding genes, eight ribosomal RNA genes, and 37 transfer RNA genes. The result of phylogenetic analysis based on 15 chloroplast genomes revealed that S. yangii is closely related to common sage (Salvia officinalis) in Lamiaceae.
Russian sage (Salvia yangii B. T. Drew), previous Perovskia atriplicifolia, was recently included in the genus Salvia (Drew et al. Citation2017). It is a flowering herbaceous perennial plant and subshrub distributed in the steppes and hills of southwestern and central Asia. Due to its adaptability to soil and climate conditions, some cultivars, such as ‘Blue Spire’, have been widely used in gardening. Additionally, as a traditional medicinal plant in its native region, S. yangii is mainly used to treat rheumatism (Majetich and Zou Citation2008). Recent studies have shown some compounds such as abrotandiol, lariciresinol, syringaresinol, and taxiresinol extracted from this plant possess the antidiabetic and anti-HBV activity (Jiang et al. Citation2015; Butt et al. Citation2019). In this study, we assembled and characterized the first complete chloroplast genome of S. yangii, which will provide organelle molecular basis for further research of this horticultural and medicinally important plant.
The whole-genome shotgun sequencing data of S. yangii leave were retrieved from sequence read archive database of NCBI (Accession: SRR6940082). The leaves of S. yangii were provided by Mountain Valley Growers, Squaw Valley, CA (GPS: 36°40′48.0″N 119°10′02.7″W; Voucher: N. García 4533, deposited at University of Florida Herbarium). In total, about 4.8 G high-quality clean reads (150 bp PE read length) were obtained with adaptors trimmed. Following Liu et al. (Citation2017, Citation2018), NOVOPlasty (Dierckxsens et al. Citation2017) was used to de novo assemble the chloroplast genome with rbcL gene as seed. Aligning and annotation were conducted using GeSeq and GENEIOUS prime (Biomatters Ltd, Auckland, New Zealand).
The complete chloroplast genome of S. yangii (GenBank accession No. MT537168) has a length of 151,473 bp with a typical circle structure and a GC content of 38.1%. It consists of a large single-copy region (LSC, 82,701 bp, 36.2% GC content), a small single-copy region (SSC, 17,566 bp, 31.9% GC content), and two inverted repeat regions (IR, 25603 bp, 43.1% GC content). In total, there were 129 genes in S. yangii, including 84 protein-coding genes, eight rRNA genes, and 37 tRNA genes. Among them, six protein-coding genes (rpl2, rpl23, ycf2, ycf15, ndhB, and rps7), seven tRNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU), and all four rRNA genes (rrn16, rrn23, rrn4.5, and rrn5) have two copies. Nine of these protein-coding genes (rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB, and ndhA) have one intron each and two (ycf3 and clpP) of them have two introns.
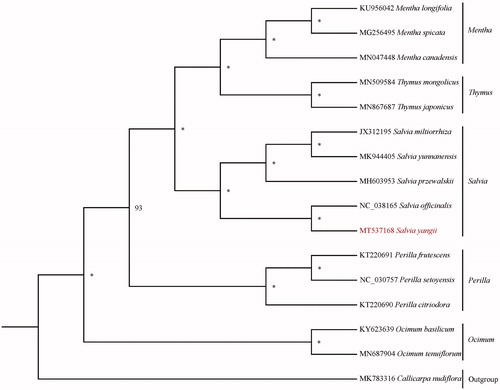
Sixteen species with available chloroplast genomes in Lamiaceae were selected to study the phylogentic placement of S. yangii (). The sequence alignment was conducted by MAFFT v1.3 (Katoh and Standley Citation2013). We drew phylogenetic tree by the software IQtree (Nguyen et al. Citation2015) with 5000 bootstrap replicates and TVM + F+R2 model. The result of phylogenetics analysis showed that S. yangii is sister to Salvia officinalis in Lamiaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The DNA matrix and phylogenetic tree that support the findings of this study are openly available in Github at https://github.com/andresqi/Russian-sage-chloroplast-genome
Additional information
Funding
References
- Butt HA, Butt HA, Khan AU. 2019. In silico analysis of compounds derived from Perovskia atriplicifolia for their antidiabetic potential. LDDD. 16(9):1074–1088.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18–e18.
- Drew BT, Gonzalez-Gallegos JG, Xiang CL, Kriebel R, Drummond CP, Walker JB, Sytsma KJ. 2017. Salvia united: the greatest good for the greatest number. Taxon. 66(1):133–145.
- Jiang ZY, Li ZQ, Huang CG, Zhou J, Hu QF, Liu WX, Huang XZ, Wang W, Zhang LZ, Xia FT. 2015. Abietane diterpenoids from Perovskia atriplicifolia and their anti-HBV activities. Bull Korean Chem Soc. 46(25):623–627.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu LX, Li P, Zhang HW, Worth J. 2018. Whole chloroplast genome sequences of the Japanese hemlocks, Tsuga diversifolia and T-sieboldii, and development of chloroplast microsatellite markers applicable to East Asian Tsuga. J Res. 23(5):318–323.
- Liu LX, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese Bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of fagales. Front Plant Sci. 8:15.
- Majetich G, Zou G. 2008. Total synthesis of (-)-barbatusol, (+)-demethylsalvicanol, (-)-brussonol, and (+)-grandione. Org Lett. 10(1):81–83.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.