Abstract
Aspergillus cristatus are the dominantly present microorganisms in dark tea. The whole mitochondrial genome sequence of A. cristatus was sequenced and reported in this study. The mitochondrial genome in A. cristatushas a full length of 77,649 bp, which is reported to be the longest among the mitochondrial genomes of Aspergillus species. The basesincluding A (34.14%), T (37.64%), C (15.61%) and G (12.61%) are found in their genome. A total of 42 genes (15 protein-coding genes, lrRNA/srRNA and 25 tRNAs) are encoded by the mitochondrial genome of this fungus. Phylogenetic analysis showed a closest relationship betweenA. pseudoglaucusand the taxonomic status of A. cristatus.
Aspergillus cristatus form the dominantly present microorganism in dark tea produced in China, these are abundant during dark tea fermentation (Xu et al. Citation2011) and are closely associated with the quality of dark tea (Wuchang et al. Citation2015). Moreover, these can also be used to treat herbal medicine residues to produce anthraquinones with medicinal value (Kong et al. Citation2019), hence, they are an important microbial resource. In addition, A. cristatus present completely different reproductive patterns under different osmotic pressures, and can be used as a good strain to study the mechanism of asexual and sexual reproduction of fungi (Ren et al. Citation2018; Yongyi et al. 2016). To date, the research on the mitochondrial genome information of A. cristatus has not been reported. Therefore, in this study, the whole mitochondrial genome sequence of A. cristatus was sequenced and reported, it was compared with the reported mitochondrial genome information of related species of fungi to further improve their taxonomic information.
The strains of A. cristatus were isolated from Fuzhuan dark tea samples which were collected in Anhua County, Hunan Province, China (N28°38′, E111°22′), the sample has been stored at Culture Collection of Hunan City University (specimen number: AC2019081217). The mitochondrial genome of A. cristatus was extracted using DNeasy Plant Mini Kit (Qiagen, Valencia, CA). Mitochondrial DNA was sequenced with the Illumina miseq 2500 platform (Illumina, San Diego, ca). Adapter and low-quality reads were deleted by NGS QC Toolkit (Patel et al. Citation2012), and the genome was annotated through MITOS web server (Bernt et al. Citation2013). Further, the annotated mitochondrial genome sequence of A. cristatus was submitted to the GenBank database (accession number: MT457782).
The mitochondrial genome of A. cristatus is a closed circular double-stranded DNA molecule comprising 77,649 bp, it is the longest among the reported mitochondrial genome of Aspergillus. The four bases of the mitochondrial genome include A (34.14%), T (37.64%), C (15.61%) and G (12.61%), comprising A + T content of 71.78%, and AT bias was obvious. The mitochondrial genome of A. cristatus encodes 42 genes, including 15 protein-coding genes, 25 tRNAs and lrRNA/srRNA, with conservative gene sequences, which is consistent with the typical characteristics of Aspergillus (Joardar et al. Citation2012).
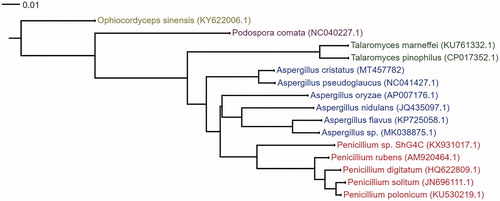
Cluster analysis of the whole mitochondrial genome sequence of 15 species of microorganisms, including A. cristatus, was performed using the neighbour-joining method. The phylogenetic tree () revealed a closest relationship between A. pseudoglaucus and the taxonomic status of A. cristatus (Park et al. Citation2019). The sequencing results can be further employed in the comparative genomics analysis of Aspergillus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT457782.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Joardar V, Abrams NF, Hostetler J, Paukstelis PJ, Pakala S, Pakala SB, Zafar N, Abolude OO, Payne G, Andrianopoulos A, et al. 2012. Sequencing of mitochondrial genomes of nine Aspergillus and Penicillium species identifies mobile introns and accessory genes as main sources of genome size variability. BMC Genomics. 13(1):698.
- Kong W, Huang C, Shi J, Li Y, Jiang X, Duan Q, Huang Y, Duan Y, Zhu X. 2019. Recycling of Chinese herb residues by endophytic and probiotic fungus Aspergillus cristatus CB10002 for the production of medicinal valuable anthraquinones. Microb Cell Fact. 18(1):102.
- Park J, Kwon W, Huang X, Mageswari A, Heo I-B, Han K-H, Hong S-B. 2019. Complete mitochondrial genome sequence of a xerophilic fungus. Aspergillus pseudoglaucus. 4(2):2422–2423.
- Patel RK, Mukesh J, Zhanjiang L. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7(2):e30619.
- Ren XX, Wang YC, Liu YX, Tan YM, Ren CG, Ge YY, Liu ZY. 2018. Comparative transcriptome analysis of the calcium signaling and expression analysis of sodium/calcium exchanger in Aspergillus cristatus. J Basic Microbiol. 58(1):76–87.
- Wuchang L, Yunlong Q, Jianan H, Lijun Y, Zhonghua L. 2015. Studies on Eurotium cristatum fungus growing affects quality tasting ingredients of primary dark tea. J Food Saf Qual. 6(5):1554–1560.
- Xu A, Wang Y, Wen J, Liu P, Liu Z, Li Z. 2011. Fungal community associated with fermentation and storage of Fuzhuan brick-tea. Int J Food Microbiol. 146(1):14–22.
- Yongyi G, Yuchen W, Yongxiang L, Yumei T, Xiuxiu R, Xinyu Z, Kevin DH, Yongfeng L, Zuoyi L. 2016. Comparative genomic and transcriptomic analyses of the Fuzhuan brick tea-fermentation fungus Aspergillus cristatus. BMC Genomics. 17:428