Abstract
The mitogenome of Eptesicus serotinus (Serotine bat) was published in 2013 with GenBank accession number KF111725 and NCBI Reference Sequence number NC_022474. This sequence was placed with Vespertilio sinensis (Asian parti-colored bat) in a COI gene tree but with Hypsugo alashanicus (Alashanian pipistrelle) in a cytochrome b gene tree. Direct comparison of mitogenomes showed that 92.4% of this mitogenome is similar to Vespertilio sinensis, 5.9% to Hypsugo alaschanicus, and that 1.6% of the mitogenome could not be attributed to either species, or any other species. This mitogenome has been re-used in at least 17 phylogenies. Our findings suggest that mitogenomes are best verified with multiple gene trees, followed by direct comparison of sequences. We conclude that greater vigilance is warranted to ensure that problematic sequences do not enter the scientific record, and are not re-used in subsequent studies.
Introduction
During the last two decades, complete mitochondrial genomes (hereafter mitogenomes) have begun to shed light on the phylogenetic relationships among bats, both at deep phylogenetic levels and among species (Nikaido et al. Citation2000; Meganathan et al. Citation2012; Botero-Castro et al. Citation2013). One of these, the mitogenome of Eptesicus serotinus (Serotine bat; KF111725/NC_022474) was released on GenBank in September 2013 and was accompanied by a descriptive paper which appeared online in July 2013 (Nam et al. Citation2015). No information was provided on its collection locality, preservation state (specimen or tissue sample only) and collection number (if any). At the time of publication, only few bat mitogenomes had become available and even today this is still the only mitogenome purportedly of this species. As a result, KF111725 or NC_022474 have been re-used in at least 17 phylogenies (Hwang et al. Citation2016; Locatelli et al. Citation2016; Monadjem et al. Citation2016; Qian et al. Citation2016; Yu et al. Citation2016; Zhang et al. Citation2016; Jebb, Foley, Kerth, et al. Citation2017; Jebb, Floey, Puechmaille, et al. Citation2017; Kim, Kim, et al. Citation2017; Kim, Park, et al. Citation2017; López-Wilchis et al. Citation2017; Mata et al. Citation2017; Shi et al. Citation2017; Platt et al. Citation2018; Huang et al. Citation2019; Yue et al. Citation2019; Tang et al. Citation2020).
In his PhD dissertation, Botero-Castro (Citation2014) noted that KF111725 is not a mitogenome of E. serotinus but represents a misidentified Vespertilio sinensis (Asian parti-colored bat). Here we show that this mitogenome is actually a chimera of V. sinensis and Hypsugo alaschanicus (Alashanian pipistrelle) and indeed does not contain any DNA fragments of E. serotinus.
Materials and methods
We verified the identity of KF111725 using sequences from two protein-coding genes: cytochrome oxidase subunit I (COI, 698 bp) and cytochrome b (cyt b, 1140 bp). These are the two most commonly used mitochondrial markers in chiropteran systematics. A phylogeny was also constructed using sequences of the 13 protein-coding genes (PCGs) included in the mitogenome but with the data set trimmed by GBLOCKS (Castresana Citation2000). GBLOCKS eliminates poorly aligned positions and divergent regions, which may not be homologous or may have been saturated by multiple substitutions (Castresana Citation2000). This resulted in an alignment of 11,328 bp. The MITOS2 web server (Bernt et al. Citation2013) was used to obtain information on the first and last positions of individual genes. CLUSTALW (as implemented in MEGA7, Kumar et al. Citation2016) was used to align sequences. Maximum Likelihood phylogenies were obtained using MEGA7. The appropriate substitution model for each data set was selected using the Akaike Information Criterion. In all cases, the GTR + G + I model was selected. Sequence divergence was calculated as uncorrected p-values with complete deletion of nucleotide positions with missing data.
Results
In the COI gene tree (), KF111725 was part of a strongly-supported clade with five sequences of V. sinensis, to which it was closely similar (0.2–0.6% sequence divergence). In contrast, in the cyt b gene tree () KF111725 was part of the Hypsugo clade, sister to four H. alashanicus from which it was 2.5–2.9% divergent. Direct comparison of aligned sequences showed that bp 1–136 of the cyt b fragment of KF111725 were a close match with V. sinensis (0–0.7% divergence), whereas bp 137–1140 were a close match with four H. alaschanicus (0.3–0.9% divergence).
Figure 1. ML phylogenies of vespertilionid bats and selected outgroups based on (a) COI (698 bp), (b) cytochrome b (1140 bp) and (c) mitogenomes (13 PCGs, trimmed with GBLOCKS; 11,328 bp). Numbers along branches represent bootstrap support values (>70%) based on 1000 pseudoreplications. Note that KF111725 (Eptesicus serotinus) clustered with Vespertilio sinensis in the COI gene tree, but with Hypsugo alashanicus in the cytochrome b gene tree.
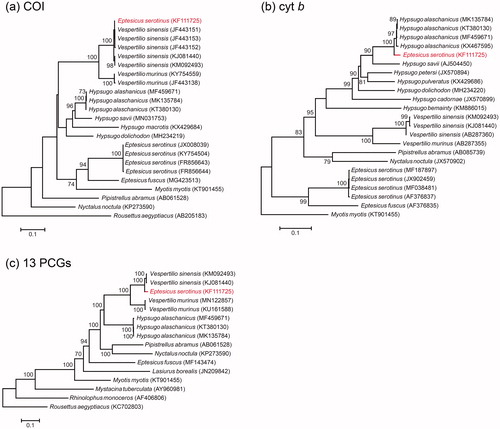
ML analysis of the GBLOCKS-trimmed 13 PCGs placed KF111725 sister to two V. sinensis () but differed from those sequences by 2.7–3.0%. Direct comparison of KF111725 with the full mitogenomes of V. sinensis (KJ081440, KM092493) and H. alaschanicus (MK135784, MF459671, KT380130) showed that 92.4% (bp 1–14,301, 15,308–15,895, 16,151–16,673) of this mitogenome is that of V. sinensis and 5.9% (bp 14,308–15,294) is of H. alaschanicus. A fragment of 243 bp (bp 15,908–16,150; 1.6% of the mitogenome) could not be attributed to either species, or indeed to any other species.
Discussion
The placement of KF111725 with V. sinensis in the COI gene tree but with H. alashanicus in the cytochrome b gene tree is prima facie evidence that this mitogenome represents a chimera. Direct comparison of KF111725 with sequences of V. sinensis and H. alaschanicus clearly show that a major part of the mitogenome is nearly identical to that of the former species, and that another part is identical to that of the latter species. This shows that KF111725 is a chimera of V. sinensis and H. alaschanicus. The laboratory that produced this mitogenome has indeed also produced mitogenomes of V. sinensis (Yoon et al. Citation2016) and H. alaschanicus (Kim and Park Citation2015; Kim et al. Citation2019). However, because none of these sequences contained any DNA of E. serotinus, we suspect that KF111725 was initially misidentified as V. sinensis, followed by transfer of a DNA fragment of H. alaschanicus, either as template DNA prior to PCR amplification, or as PCR product prior to sequencing, or as a DNA sequence fragment during sequence assembly/editing. In any case, the problematic nature of KF111725 (and NC_022474) shows that this mitogenome should not be used for any evolutionary applications.
To enable efficient detection of misidentified mitogenomes, Botero-Castro et al. (Citation2016) proposed that mitogenome announcements should include (i) a ML gene tree of the most commonly used barcode marker in the group be used to assess the identity of the mitogenome, and (ii) a ML tree of mitogenomes of closely related species, and that (iii) relationships be depicted with phylograms to show branch lengths. Whilst we agree with these recommendations, our study shows that these may not be sufficient for diagnosing chimeras. Whereas both the COI and mitogenome trees clearly show that KF111725 is not a mitogenome of E. serotinus, the placement of KF111725 in a clade with V. sinensis could also be explained by misidentification. Similarly, if only the cyt b gene tree were used to verify the identity of KF111725, its placement close to, but distinct from, sequences of H. alashanicus in itself would not be evidence of a chimera and could also be explained by it being a representative of a previously undocumented population of that species. The discovery of novel lineages within a wide-ranging species is a common finding in bat phylogeography (e.g., Larsen et al. Citation2012; Juste et al. Citation2013; Tu et al. Citation2018). Only when either (i) both COI and cyt b gene trees are analyzed, or (ii) cyt b sequences are directly compared with those of V. sinensis and H. alaschanicus, would it become evident that KF111725 is not a misidentified sequence. In addition, neither in the two single gene trees nor in the mitogenome tree KF111725 was placed on an unusually long branch, which is probably another factor that made it difficult to detect and diagnose this sequence as a chimera. Finally, the lack of a bona fide mitogenome of E. serotinus, and the late publication of a mitogenome of another member of the genus (Big brown bat E. fuscus; Platt et al. Citation2018), likely also contributed to the late diagnosis of this mitogenome as a chimera. We recommend that mitogenomes are best verified with multiple gene trees, and that any potentially problematic mitogenomes are directly compared with those of all relevant species or genera.
Assessment of the consequences of re-usage of KF111725 and NC_022474 in other papers is outside the scope of this paper. However, we note that this mitogenome sequence has already been used (i) for phylogeny reconstruction, either as ingroup (e.g. Qian et al. Citation2016; Yu et al. Citation2016; Zhang et al. Citation2016; López-Wilchis et al. Citation2017) or outgroup (e.g. Platt et al. Citation2018), (ii) for primer design (Yoon et al. Citation2016), and (iii) in a reference database for DNA screening of Finnish stone age sediments (Peltola Citation2019). We conclude that greater vigilance is warranted to ensure that problematic sequences do not enter the scientific record, and that those that remain undetected do not compromise evolutionary inferences.
Disclosure statement
No potential conflict of interest was reported by the author(s)
Data availability statement
The data that support the findings of this study are openly available on GenBank at https://www.ncbi.nlm.nih.gov/nucleotide. Accession numbers are listed in .
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Botero-Castro F. 2014. Systématique, phylogénie et évolution moléculaires des Phyllostomidae (Mammalia, Chiroptera): une approche mitogénomique comparative [PhD dissertation]. Biologie moléculaire. Université Montpellier II – Sciences et Techniques du Languedoc; [accessed 2020 Feb 26]. https://tel.archives-ouvertes.fr/tel-01344308/document.
- Botero-Castro F, Delsuc F, Douzery EJ. 2016. Thrice better than once: quality control guidelines to validate new mitogenomes. Mitochondrial DNA Part A. 27(1):449–454.
- Botero-Castro F, Tilak MK, Justy F, Catzeflis F, Delsuc F, Douzery EJ. 2013. Next-generation sequencing and phylogenetic signal of complete mitochondrial genomes for resolving the evolutionary history of leaf-nosed bats (Phyllostomidae). Mol Phylogenet Evol. 69(3):728–739.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Huang Z, Yue Y, Thapa S, Hu Y, Wu Y, Yu W. 2019. Mitochondrial genome of Murina shuipuensis (Chiroptera: Vespertilionidae) from Shuifu Village, Guizhou, China (type locality. Mitochondrial DNA Part B. 4(2):2588–2590.
- Hwang JY, Jin GD, Park J, Lee SG, Kim EB. 2016. Complete sequences of eastern water bat, Myotis petax (Chiroptera; Microchiroptera; Vespertilionidae) mitogenome. Mitochondrial DNA Part A. 27(5):3715–3716.
- Jebb D, Foley NM, Kerth G, Teeling EC. 2017. The complete mitochondrial genome of the Bechstein’s bat, Myotis bechsteinii (Chiroptera, Vespertilionidae). Mitochondrial DNA Part B. 2(1):92–94.
- Jebb D, Foley NM, Puechmaille SJ, Teeling EC. 2017. The complete mitochondrial genome of the Greater mouse-eared bat, Myotis myotis (Chiroptera: Vespertilionidae). Mitochondrial DNA Part A. 28(3):347–349.
- Juste J, Benda P, Garcia‐Mudarra JL, Ibanez C. 2013. Phylogeny and systematics of Old World serotine bats (genus Eptesicus, Vespertilionidae, Chiroptera): an integrative approach. Zool Scr. 42(5):441–457.
- Kim JY, Kim HR, Lim SJ, Kim HJ, Cho JY, Park YC. 2017. Complete mitochondrial genome of the house bat Pipistrellus abramus (Mammalia: Chiroptera) from Korea. Mitochondrial DNA Part B. 2(2):540–541.
- Kim JY, Park YC. 2015. Gene organization and characterization of the complete mitogenome of Hypsugo alaschanicus (Chiroptera: Vespertilionidae). Genet Mol Res. 14(4):16325–16331.
- Kim KY, Yoon KB, Park YC. 2019. Phylogenetic position of the Hypsugo alaschanicus based on complete mitochondrial genome sequences. Mitochondrial DNA Part B. 4(1):620–621.
- Kim YK, Park SG, Kim TW, Park JH, Adhikari P, Kim G, Park SM, Lee JW, Oh DJ, Han SH, Oh HS. 2017. Complete mitochondrial genome of the far eastern Myotis, Myotis bombinus (Chiroptera, Vespertilionidae). Mitochondrial DNA Part A. 28(2):267–268.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Larsen RJ, Knapp MC, Genoways HH, Khan FAA, Larsen PA, Wilson DE, Baker RJ. 2012. Genetic diversity of neotropical Myotis (chiroptera: vespertilionidae) with an emphasis on South American species. PLOS One. 7(10):e46578.
- Locatelli AG, Jebb D, Teeling EC. 2016. The complete mitochondrial genome of Kuhl’s pipistrelle, Pipistrellus kuhlii (Chiroptera: Vespertilionidae). Mitochondrial DNA Part B. 1(1):423–424.
- López-Wilchis R, Del Río-Portilla MÁ, Guevara-Chumacero LM. 2017. Mitochondrial genome of Pteronotus personatus (Chiroptera: Mormoopidae): comparison with selected bats and phylogenetic considerations. Genetica. 145(1):27–35.
- Mata VA, Amorim F, Guillén-Servent A, Beja P, Rebelo H. 2017. First complete mitochondrial genomes of molossid bats (Chiroptera: Molossidae). Mitochondrial DNA Part B. 2(1):152–154.
- Meganathan PR, Pagan HJ, McCulloch ES, Stevens RD, Ray DA. 2012. Complete mitochondrial genome sequences of three bats species and whole genome mitochondrial analyses reveal patterns of codon bias and lend support to a basal split in Chiroptera. Gene. 492(1):121–129.
- Monadjem A, Joubert C, Richards L, Nielsen IB, Nielsen M, Kjartansdóttir KR, Bohmann K, Mourier T, Hansen AJ. 2016. First record of Vespertilio murinus from the Arabian Peninsula. Vespertilio. 18:79–89.
- Nam TW, Yoon KB, Cho JY, Park YC. 2015. Complete mitochondrial genome of the serotine bat (Eptesicus serotinus) in Korea. Mitochondrial DNA. 26(3):459–460.
- Nikaido M, Harada M, Cao Y, Hasegawa M, Okada N. 2000. Monophyletic origin of the order Chiroptera and its phylogenetic position among Mammalia, as inferred from the complete sequence of the mitochondrial DNA of a Japanese megabat, the Ryukyu flying fox (Pteropus dasymallus). J Mol Evol. 51(4):318–328.
- Peltola S. 2019. Ancient DNA screening from Finnish stone age sediments [MSc Thesis]. University of Helsinki; [accessed 2020 Mar 2]. https://helda.helsinki.fi/bitstream/handle/10138/306560/Peltola_Sanni_pro_gradu_2019.pdf?sequence=2.
- Platt RN, Faircloth BC, Sullivan KAM, Kieran T, Glenn TC, Vandewege MW, Lee TE, Baker RJ, Stevens RD, Ray DA. 2018. Conflicting evolutionary histories of the mitochondrial and nuclear genomes in New World Myotis Bats. Syst Biol. 67(2):236–249.
- Qian K, Yu D, Cheng H, Storey KB, Zhang J. 2016. The complete mitochondrial genome of Nyctalus noctula (Chiroptera: Vespertilionidae). Mitochondrial DNA Part A. 27(4):2365–2366.
- Shi Y, Zhao S, Han X, Xu C. 2017. Revealing the complete mitogenome sequence of Hypsugo alaschanicus based on next generation sequencing. Mitochondrial DNA Part B. 2(2):575–576.
- Tang X, Yu W, Wu Y. 2020. Mitochondrial genome of the Harpiocephalus harpia (Chiroptera: Vespertilionidae) from China. Mitochondrial DNA Part B. 5(1):996–998.
- Tu VT, Hassanin A, Furey NM, Son NT, Csorba G. 2018. Four species in one: multigene analyses reveal phylogenetic patterns within Hardwicke's woolly bat, Kerivoula hardwickii-complex (Chiroptera, Vespertilionidae) in Asia. Hystrix. 29:111–121.
- Yoon KB, Lee JH, Cho JY, Park YC. 2016. The complete mitochondrial genome of the Asian particolored bat Vespertilio sinensis (Chiroptera: Vespertilionidae) in Korea. Mitochondrial DNA Part A. 27(1):299–300.
- Yu D, Qian K, Storey KB, Hu Y, Zhang J. 2016. The complete mitochondrial genome of Myotis lucifugus (Chiroptera: Vespertilionidae). Mitochondrial DNA Part A. 27(4):2423–2424.
- Yue Y, Huang Z, Li F, Thapa S, Hu Y, Wu Y, Yu W. 2019. The complete mitochondrial genome of the tube-nosed bat Murina cyclotis (Chiroptera: Vespertilionidae) in China. Mitochondrial DNA Part B. 4(2):2248–2250.
- Zhang Q, Cong H, Yu W, Kong L, Wang Y, Li Y, Wu Y. 2016. The mitochondrial genome of Murina huttoni rubella (Chiroptera: Vespertilionidae) from China. Mitochondrial DNA Part B. 1(1):438–440.
