Abstract
Lepidium meyenii Walp. is a frequently used medicinal plant (namely, ‘maca’) in Yunnan Province of China. In this study, we sequenced the complete chloroplast (cp) genome sequence of L. meyenii to investigate its phylogenetic relationship in the family Cruciferae. The chloroplast genome of L. meyenii was 154,839 bp in length with 36.39% overall GC content, including a large single copy (LSC) region of 83,943 bp, a small single copy (SSC) region of 17,978 bp and a pair of inverted repeats (IRs) of 52,918 bp. The cp genome contained 103 genes, including 78 protein-coding genes, 21 tRNA genes, and four rRNA genes. The phylogenetic analysis indicated L. meyenii was closely related to the genus Capsella.
Lepidium meyenii is a 1-year or biennial herb of the Lepidium genus in the cruciferous family. It is native to the Andean region of South America with low temperatures, strong winds and harsh ecological conditions at an altitude of 3500 − 4500 m. Local people have a long history of planting and eating maca, and there are reports in the literature that they have been eating it for more than 5800 years (Li et al. Citation2018). There is no history of eating maca in our country, however, after the medicinal plant was included in the new food resource catalog in May 2011, the consumption of maca in China has a legal basis (Li et al. Citation2018). Lijiang and huize of yunnan are important producing areas for introducing L. meyenii in China. The main edible part of maca is its rhizome, also known as beetroot or Peruvian ginseng. The main edible part of L. meyenii is its rhizome, also known as beetroot or Peruvian ginseng, which has a long history as medicine and food, rich in nutrition and remarkable efficacy. Pharmacological activity studies show that L. meyenii has a variety of effects such as anti-fatigue, enhance immunity, improve fertility, anti-inflammatory, anti-cancer, antioxidant, anti-viral, lower blood pressure, relieve depression and treat women's menopause syndrome (Wang et al. Citation2007). However, until now, most of the studies for this species mainly focused on describing its chemical compositions (Zhou et al. Citation2016; Zhou Citation2017) with little involvement in its molecular biology. Here, we reported the complete chloroplast genome sequence of L. meyenii and revealed its phylogenetic relationships with other species in the Cruciferae.
Fresh and clean leaf materials of L. meyenii were sampled from Huize County, Yunnan, China (N26.41°, E103.27°); meanwhile, a voucher specimen (No. ZHZ001) was collected and deposited at the Herbarium of Medicinal Plants and Crude Drugs of the College of Pharmacy and Chemistry, Dali University. The total genomic DNA was extracted using the improved CTAB method (Doyle Citation1987; Yang et al. Citation2014), and sequenced with Illumina Hiseq 2500 (Novogene, Tianjing, China) platform with pair-end (2 × 300 bp) library. About 4.11 Gb of raw reads with 20,623,534 paired-end reads were obtained from high-throughput sequencing. The raw data was filtered using Trimmomatic v.0.32 with default settings (Bolger et al. Citation2014) were assembled into circular contigs using GetOrganelle.py (Jin et al. Citation2018). Finally, the cpDNA was annotated by the Dual Organellar Genome Annotator (DOGMA; http://dogma.ccbb.utexas.edu/) (Wyman et al. Citation2004) and tRNAscan-SE (Lowe and Chan Citation2016).
The annotated chloroplast genome was submitted to the GenBank with an accession number MT430983. The total length of the chloroplast genome was 154,839 bp, with 36.39% overall GC content. With typical quadripartite structure, a pair of IRs (inverted repeats) of 52,918 bp was separated by a small single copy (SSC) region of 17,978 bp and a large single copy (LSC) region of 83,943 bp. The cp genome contained 130 genes, including 78 protein-coding genes, 21 tRNA genes, and four rRNA genes. Of these, 17 genes were duplicated in the inverted repeat regions, 16 genes, and 6 tRNA genes contain one intron, while two genes (ycf3 and clpP) have two introns.
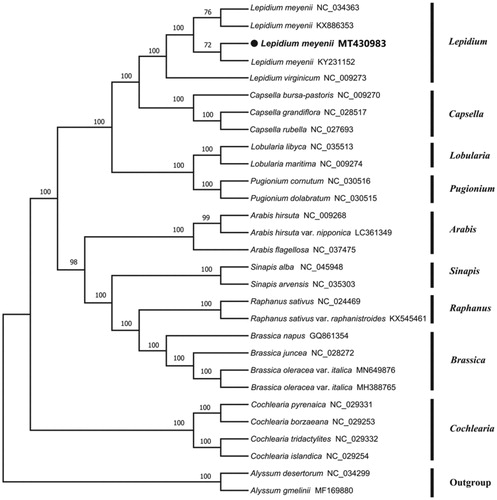
To investigate its taxonomic status, a total of 28 cp genome sequences of Cruciferae species were downloaded from the NCBI database used for phylogenetic analysis. After using MAFFT V.7.149 for aligning (Katoh and Standley Citation2013), a neighbor-joining (NJ) tree was constructed in MEGA v.7.0.26 (Kumar et al. Citation2016) with 1000 bootstrap replicates and two Cruciferae species (Alyssum desertorum:NC_034299 and Alyssum gmelinii: MF169880) were used as outgroups. The results showed that L. meyenii was closely related to the genus Capsella (). Meanwhile, the phylogenetic relationship in Cruciferae was consistent with previous studies and this will be useful data for developing markers for further studies.
Disclosure statement
The authors are highly grateful to the published genome data in the public database. The authors declare no conflicts of interest and are responsible for the content.
Data Availability Statement
Date available. Please access the GenBank and obtain it in https://www.ncbi.nlm.nih.gov/nuccore/MT430983.
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv.DOI:10.1101/256479
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li AM, Wang C, Zhang JJ, Wang LY, Gao XD, Huang YG. 2018. Research and analysis on the current situation of maca. Chin J Tradit Chin Med Pharm. 33(01):244–248.
- Li Y, Li PY, Zhou XT, Zhou LY, Huang LQ, Yang G, Chen M. 2018. [Research and application progress of Lepidium meyenii (maca)]. Zhongguo Zhong Yao Za Zhi. 43(23):4599–4607.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Wang Y, Wang Y, McNeil B, Harvey LM. 2007. Maca: an andeancrop with multi-pharmacological functions. Food Res Int. 40(7):783–792.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031.
- Zhou Y, Luan J, Chu ZY. 2016. Advances in the study of chemical constituents and their pharmacological effects of maca. J Changsha Med Univ.
- Zhou YY. 2017. Chemical analysis of Maca and the mechanism of the neuroprotective effect on its active ingredients. Beijing: China Academy of Chinese Medical Sciences, PhD.