Abstract
We report the complete mitochondrial genome of the slender racer (Orientocoluber spinalis), which is widely distributed in Northeast Asia, using a specimen collected from Mt. Worak, South Korea. The mitogenome was 17,196 bp long, consisting of 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and 3 noncoding regions. The presented sequence could be useful for evolutionary and phylogenetic studies of Colubridae.
Orientocoluber (formerly Hierophis) spinalis, called the slender racer, is the only species in the genus Orientocoluber (subfamily Colubrinae), which was newly coined in 2011 (Kharin Citation2011). The species has mainly been found in midcentral and eastern China and most areas of North and South Korea and has occasionally been reported in eastern Kazakhstan, Mongolia, and Far East Russia (Hoser Citation2012; Maslova et al. Citation2018). Due to its cryptic lifestyle and fast movements (Kim Citation2011), no phylogenetic or ecological studies have been conducted on this species. In this study, we report the complete mitochondrial genome sequence of O. spinalis, which can be used for future phylogenetic and evolutionary studies of O. spinalis and closely related species.
We obtained a complete mitogenome sequence for O. spinalis, which was collected from Mt. Worak, South Korea (36.80243° and 128.05404°E), in 2019. The specimen was preserved in 95% ethanol and kept in the herpetology laboratory of Kangwon National University (Voucher No. G01000OS). We extracted whole-genomic DNA from the muscle tissue of the specimen using Qiagen’s DNeasy Blood and Tissue Kit (Qiagen Korea Ltd., Seoul, South Korea) and sequenced it using the MiSeq platform (Illumina, San Diego, CA). To assemble and annotate the full mitochondrial DNA sequence, we used Geneious version 9.0.4 (Biomatters Ltd., Auckland, New Zealand), tRNA Scan-SE1.21 software (http://lowelab.ucsc.edu/tRNA Scan-SE/) (Lowe and Eddy Citation1997), and MITOS (http://mitos2.bioinf.uni-leipzig.de/) (Bernt et al. Citation2013). The mitogenome of O. spinalis was annotated based on the mitogenomes of five species in subfamily Colubrinae (GenBank Nos. KF148620, KF148621, KF750656, KJ719252, and KP888955). We first determined the phylogenetic position of our specimen using partial cytochrome b (Cytb, 1117 bp) genes from our specimen, O. spinalis (GenBank No. AY376744), and 16 other colubrid snakes (GenBank Nos. AF471049, AF471054, AY188025-6, AY188043, AY376741, AY376747, AY376749, AY376759, AY376760, AY486915, AY486917, AY486933, DQ902108, and DQ902128). Subsequently, we constructed a phylogenetic tree of O. spinalis and 16 snakes from 3 families (Colubridae, Elapidae, and Viperidae) using the complete mitogenome sequence obtained in this study and complete mitogenome sequences from GenBank () with PAUP* version 4.0b10 (Swofford Citation2003). We applied Bayesian inference (BI) and maximum likelihood (ML) methods to construct the tree using 1000 bootstrap replicates.
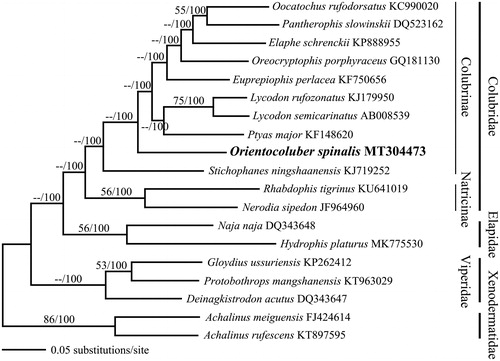
Figure 1. BI tree of Orientocoluber spinalis with 16 species from 3 families (Colubridae, Elapidae, and Viperidae) based on its complete mitogenome. Two species from family Xenodermatidae were used as the outgroup. Complete mitogenome sequences were downloaded from GenBank, with the accession number indicated after the scientific name of each species. On each branch, ML bootstrap value/Bayesian posterior probabilities are denoted.
The mitogenome length of O. spinalis was 17,196 bp (GenBank Accession No. MT304473), and its base composition was 34.0% A, 25.2% T, 13.0% G, and 27.8% C, revealing an A-T rich (59.2%) composition. The mitogenome contained 13 protein-coding genes, 2 rRNA genes (12S and 16S rRNA), 22 tRNA genes, and 3 non-coding regions, namely, two displacement loop regions and an L-strand replication origin. The arrangement and transcription directions of the mitogenome were identical to those of other species in subfamily Colubrinae (Ptyas dhumnades [GenBank No. KF148621] and Stichophanes ningshaanensis [GenBank No. KJ719252]).
In the phylogenetic tree of partial Cytb gene sequences, our specimen was placed within the lineage of existing O. spinalis, compared to other closely related species, as shown in Pyron et al. (Citation2011). This placement confirmed that our specimen was O. spinalis. In the phylogenetic tree of the mitogenome, O. spinalis formed an independent lineage compared with the other Colubrinae species, similar to the result of Zheng and Wiens (Citation2016), who constructed a phylogeny for 4162 squamate taxa using 52 different genes. Our results could be important for further understanding the phylogenetic evolution of Colubridae.
Geolocation information
Mt. Worak in 61-3, Songgye-ri, Hansu-myeon, Jecheon-si, Chungcheongbuk-do, South Korea (36°80‘24.3“N and 128°05‘40.4“E).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in ‘figshare’ at http://doi.org/10.6084/m9.figshare.12284402
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Hoser RT. 2012. Yeomansus: a new genus for the slender racer (Serpentes: Colubridae). Aust J Herpetol. 14:3–5.
- Kharin VE. 2011. Rare and little-known snakes of the north-eastern Eurasia. 3. On the taxonomic status of the slender racer Hierophis spinalis (Serpentes: Colubridae). Mod Herpetol. 11:173–179.
- Kim JC. 2011. Red data book of endangered amphibians and reptiles in Korea. Incheon, Korea: National Institute of Biological Resources.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Maslova IV, Portnyagina EY, Sokolova DA, Vorobieva PA, Akulenko MV, Portnyagin AS, Somov AA. 2018. Distribution of rare and endangered amphibians and reptiles in Primorsky Krai (Far East, Russia). Nat Con Res. 3:61–72.
- Pyron RA, Burbrink FT, Colli GR, De Oca ANM, Vitt LJ, Kuczynski CA, Wiens JJ. 2011. The phylogeny of advanced snakes (Colubroidea), with discovery of a new subfamily and comparison of support methods for likelihood trees. Mol Phylogenet Evol. 58(2):329–342.
- Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony, version 4.0 b10. Sunderland (MA): Sinauer Associates.
- Zheng Y, Wiens JJ. 2016. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol Phylogenet Evol. 94(Pt B):537–547.
