Abstract
In this study, the complete mitogenome of an entomopathogenic fungus Orbiocrella petchii (syn. Torrubiella petchii) was assembled and annotated. This circular mitogenome was 23,794 bp in length and consisted of 2 rRNA genes (rnl and rns), 25 tRNA genes, and 14 standard protein-coding genes of the oxidative phosphorylation system. Two group I introns were identified, and they encoded ribosomal protein S3 (in rnl) or a GIY-YIG endonuclease (in nad1). Phylogenetic analysis based on mitochondrial DNA sequences confirms O. petchii in the family of Clavicipitaceae.
Torrubiella is characterized as arthropod pathogens that produce superficial perithecia on a loose mat of hyphae or a highly reduced non-stipitate stroma. Torrubiella petchii (currently Orbiocrella petchii) is a pathogen of scale insects found on bamboo leaves (Hywel-Jones Citation1997). This fungus was recently transferred to Orbiocrella (Johnson et al. Citation2009), which is currently a monospecific genus. This species has been shown to be a unique source of bioactive aromatic polyketides (Isaka et al. Citation2019); however, its studies at the molecular level is still very limited. Herein, we present the mitogenome of O. petchii SD3, which was isolated from a scale insect (Hemiptera) underside of a bamboo leaf in Ding Hu Mountain Nature Reserve, Guangdong Province, China (23°10′N, 112°31′E) and was deposited in the China General Microbiological Culture Collection Center (CGMCC), China (Accession 3.17637).
Total DNA of SD3 was randomly sheared to fragments of 400 bp followed by sequencing on an Illumina HiSeq 2500 platform in 2 × 250 bp reads. Mitogenome was de novo assembled from clean reads using NOVOPlasty (Dierckxsens et al. Citation2017) and then annotated as described previously (Zhang et al. Citation2017). Introns are named according to proposals suggested by Johansen and Haugen (Citation2001) and Zhang and Zhang (Citation2019).
The mitogenome of O. petchii SD3 (GenBank accession: MT447058) is a circular molecule of 23,794 bp with an AT content of 71.88%. This mitogenome encodes 2 ribosomal RNAs (rnl and rns), 25 tRNAs, and 14 conserved proteins of the oxidative phosphorylation system (nad1-6, 4 L; cob; cox1-3, and atp6, 8, 9). These tRNA genes code for all 20 standard amino acids. There are three tRNA genes for methionine with the same anticodon, two tRNA genes for arginine, leucine, and serine with different anticodons. The majority of tRNA genes are clustered upstream (trnV, I, S2, W, P) and downstream (trnT, E, M1, M2, L1, A, F, K, L2, Q, H, M3) of the rnl gene, and downstream (trnY, D, S1, and N) of the rns gene. No intergenic free-standing open reading frame (ORF) is identified. For the two neighboring gene pairs commonly found in hypocrealean species, nad3 follows immediately nad2, whereas nad5 overlaps one nucleotide with its upstream gene nad4L. It should be noted that nad6 overlaps 50 bp at the 3′ end with trnV. All protein-coding genes in the mitogenome start by ATG and terminate by TAA except cox1 which terminates by TAG. This mitogenome is rather compact with genic regions (21,793 bp) accounting for 91.5% of the mitogenome. All genes are transcribed at the same strand.
Only two introns are identified, and they invade nad1 and rnl. Both introns belong to the group I intron family but two specific subgroups, namely IB (nad1P636) and IA (mL2450). Intronic ORFs encode GIY-YIG endonuclease (in nad1) or ribosomal protein S3 (in rnl). Intronic regions (including intronic ORFs) have a total length of 2306 bp.
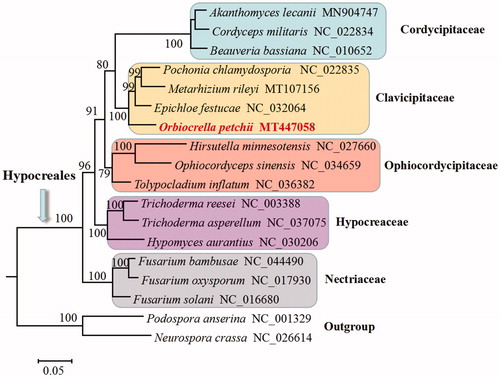
Phylogenetic analysis based on mitochondrial DNA sequences confirms O. petchii as a member of Clavicipitaceae, and it has a close relationship to the grass symbiont Epichloe festucae (). This study supports the conclusion that Torrubiella is not monophyletic due to the fact that Torrubiella petchii (currently O. petchii) clusters in Clavicipitaceae and Torrubiella confragosa (currently Akanthomyces lecanii) clusters in Cordycipitaceae (Zhang et al. Citation2020).
Figure 1. Phylogenetic analysis of Hypocreales species based on mitochondrial nucleotide sequences. We used three representative species of all families with available mitogenomes in Hypocreales. Two Sordariales species (Podospora anserine and Neurospora crassa) were used as outgroups. The whole mitogenome sequences (or exonic sequences in cases with alignment difficulties) of these species were aligned and trimmed using the HomBlocks pipeline (Bi et al. Citation2018), resulting in an alignment of 6667 characters. Phylogenetic reconstruction was performed using the maximum likelihood approach as implemented in RAxML version 8.2.12 (Stamatakis Citation2014). Support values were given for nodes that received bootstrap values ≥70%. GenBank accession numbers followed after fungal taxon names.
Acknowledgments
We thank the High Performance Simulation Platform of Shanxi University for providing computing resource.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The O. petchii SD3 mitogenome sequence was deposited in GenBank under accession number MT447058 (https://www.ncbi.nlm.nih.gov/nuccore/MT447058).
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Hywel-Jones NL. 1997. Torrubiella petchii, a new species of scale insect pathogen from Thailand. Mycol Res. 101(2):143–145.
- Isaka M, Palasarn S, Choowong W, Kawashima K, Mori S, Mongkolsamrit S, Thanakitpipattana D. 2019. Benzophenone and chromone derivatives and their dimers from the scale-insect pathogenic fungus Orbiocrella petchii BCC 51377. Tetrahedron. 75(45):130646.
- Johansen S, Haugen P. 2001. A new nomenclature of group I introns in ribosomal DNA. RNA. 7(7):935–936.
- Johnson D, Sung GH, Hywel-Jones NL, Luangsa-Ard JJ, Bischoff JF, Kepler RM, Spatafora JW. 2009. Systematics and evolution of the genus Torrubiella (Hypocreales, Ascomycota). Mycol Res. 113(Pt 3):279–289.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zhang S, Zhang Y-J. 2019. Proposal of a new nomenclature for introns in protein-coding genes in fungal mitogenomes. IMA Fungus. 10(1):15.
- Zhang Y-J, Yang X-B, Zhang S. 2020. Complete mitogenome of the entomopathogenic fungus Akanthomyces lecanii. Mitochondrial DNA Part B. 5(1):1021–1022.
- Zhang Y-J, Yang X-Q, Zhang S, Humber RA, Xu J. 2017. Genomic analyses reveal low mitochondrial and high nuclear diversity in the cyclosporin-producing fungus Tolypocladium inflatum. Appl Microbiol Biotechnol. 101(23–24):8517–8531.
