Abstract
The molecular information of lesser-known bullfrog, Hoplobatrachus litoralis is restricted from Bangladesh (type locality) and Myanmar. The morphological observation further evidenced the range-extension of this species from India and Myanmar. Here, we collected the H. litoralis specimen from the Dampa Tiger Reserve, Mizoram state and provides a new altitude record (268 m) and range-extension in northeast India. The DNA barcoding with mitochondrial Cytochrome b gene discriminates H. litoralis from other congeners with sufficient genetic distance (13.9% to 27.8%). The Bayesian phylogeny revealed monophyletic clustering of all five Hoplobatrachus species and showed the sister relationship of H. litoralis and H. tigerinus. The generated sequences of H. litoralis from northeast India depicted shallow divergence (1.5%) from the sequences generated from Bangladesh, indicates the possible genetic variability due to the alteration of the ecological niche. We suggested further physiological and molecular study with extensive sampling of this species to confirm their actual range distribution, high-altitude adaptation, and gene flow.
Introduction
The genus Hoplobatrachus (Family Dicroglossidae) consists of five species globally and widely distributed in Sub-Saharan Africa and southern and southeastern Asia (Frost Citation2020). Among them, two species Hoplobatrachus crassus and Hoplobatrachus tigerinus are antecedently known to be distributed in India. Except Hoplobatrachus litoralis, all other species are bearing Least Concern status in the International Union for the Conservation of Nature (IUCN) Red List of Threatened species. However, they are confronted the illegitimate threat of hunting and trapping for consumption as food and medicine in Asian countries (IUCN 2020). Nevertheless, owing to the rapid growth of urbanization; their suitable habitats are largely shrinking in the wild. Among all extant congeners, H. litoralis is lately described species from the Cox’s Bazar district, coastal belt of Bangladesh (Hasan et al. Citation2012). Later on, multiple specimens from different geographical locations further extended the range distribution of H. litoralis beyond its type locality. Based on the morphological observation the species was recorded from three different states (Assam, Tripura and West Bengal) in eastern and northeastern India (Purkayastha and Basak Citation2018; Mondal et al. Citation2018; Bohra et al. Citation2019) (). Besides the taxonomic studies, the molecular data of H. litoralis was also generated from Bangladesh and Myanmar (Hasan et al. Citation2012; Sultana et al. Citation2017; Mulcahy et al. Citation2018). The morphological characters often create confusion to identify the H. litoralis from their close relative H. tigerinus in different life stages. In this instance, the utility of mitochondrial genes (16S rRNA and Cytb) were evidenced to identify the species accurately and effectively discriminated H. litoralis from the sister species H. tigerinus (Mulcahy et al. Citation2018).
Figure 1. (A) Map showing the updated distribution of H. litoralis with the new altitude record and range-extension in northeast India. (B) Live photograph of H. litoralis collected from Mizoram state in northeast India. (C) Bayesian phylogeny based on partial mtCytb gene inferred the relationship of H. litoralis with other Hoplobatrachus congeners.
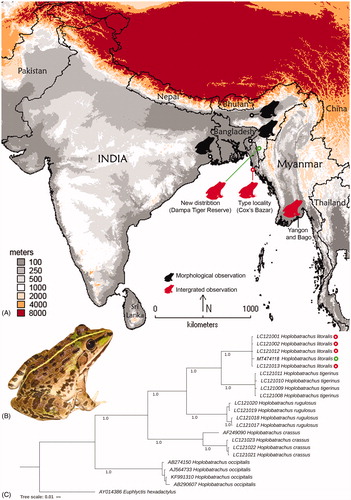
The integrated approach in combination with morphology, molecular, bioacoustics, and zoogeography is manifested to be best practice in anurans systematic studies (Padial et al. Citation2010; Ortega-Andrade et al. Citation2015; Köhler et al. Citation2017). The partial or complete mitochondrial genes were evidenced to discriminate variety of animals including anurans (Verma and Singh Citation2002; Hebert et al. Citation2003; Biju et al. Citation2014; Chambers and Hebert Citation2016; Kundu et al. Citation2020). Apart from the accurate species level identification, this molecular tool is also utilized to recognize the phylogenetic and evolutionary relationship (Biju and Bossuyt Citation2009; Van Bocxlaer et al. Citation2012; Mahony et al. Citation2017), cryptic diversity (Bickford et al. Citation2007; Tyagi et al. Citation2017), and origin as well as diversification of amphibians (Van Bocxlaer et al. Citation2006; Roelants et al. Citation2007; Garg and Biju Citation2019). Although the previous morphological studies witnessed the presence of H. litoralis in India, their genetic information was lacking from these regions. Considering the young adult specimen and lack of bioacoustics information, the present study aimed to generate the DNA sequences of this lesser-known bullfrog to affirm the morphology-based identification. This is the first molecular information of this species from India contributed to the global database for future reference.
Materials and methods
During the recent survey of herpetofauna, a single young adult female specimen of dicroglossid frog was opportunistically collected from the Teirei Range in the Dampa Tiger Reserve (23.68 N 92.45 E), Mizoram state in northeast India (). Based on the morphological characters following the original description, the species was preliminarily identified as H. litoralis (Hasan et al. Citation2012). The survey and sampling were executed after acquired the prior permission (No.A.33011/2/99-CWLW/225) from the Chief Wildlife Warden of Environment, Forests and Climate Change, Govt. of Mizoram, India. The specimen was vouchered (MZMU 1383) in the museum of Department of Zoology, Mizoram University, India. The tissue sample was collected and preserved in 70% ethanol for further molecular investigation.
The genomic DNA was extracted by using QIAamp DNA Mini Kit (Qiagen, Valencia, CA) with manufacturer’s protocols. The genomic DNA was stored at −30 °C in the Center for DNA Taxonomy laboratory, Molecular Systematics Division, Zoological Survey of India, Kolkata. The published primer pair (Verma and Singh Citation2002): mcb 398: 5′-TACCATGAGGACAAATATCATTCTG-3′ and mcb 869 5′-CCTCCTAGTTTGTTAGGGATTGATCG-3′ was used to amplify the partial mitochondrial cytochrome b (Cytb) gene on a Veriti® Thermal Cycler (Applied Biosystems, Foster City, CA). The 30 µl PCR mixture contains 10 pmol of each primer, 100 ng of DNA template, 1 × PCR buffer, 1.0–1.5 mM of MgCl2, 0.25 mM of each dNTPs, and 1 U of Taq polymerase (Takara BIO Inc., Japan). The thermal profile was set as 95 °C for 10 min; 35 cycles of 95 °C for 45 s, 51 °C for 1 min and 72 °C for 2 min and subsequent hold at 4 °C. The PCR amplified product was illustrated in 1% agarose gel containing Ethidium bromide (10 mg/ml). The PCR product was further purified using QIAquickR Gel extraction Kit (Qiagen, Valencia, CA). The cycle sequencing of the purified PCR product (15 ng) was performed with BigDye®Terminator ver. 3.1 Cycle Sequencing Kit (Applied Bio systems, Foster City, CA) by using 3.2 Picomoles of PCR primer pair. The thermal profile of the Cycle Sequencing was set as 96 °C for 1 min, followed by 25 cycles of 96 °C for 10 s, 50 °C for 5 s and a final extension at 60 °C for 1 min 15 s on a Veriti® Thermal Cycler. The cycle sequencing product was cleaned by BigDye X-terminator kit (Applied Biosystems, Foster City, CA) and Sanger sequencing was performed on 48 capillary ABI 3730 Genetic analyzer housed at Zoological Survey of India, Kolkata.
The generated sequence was checked by SeqScanner V1.0 (Applied Biosystems Inc., CA, USA), nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/), and ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) to avoid the low quality reads and gaps. The generated sequence was submitted to the GenBank to acquire the accession number. Further, a total of 20 publicly available database sequences of Hoplobatrachus species were acquired from GenBank to form a combined dataset to estimate the genetic distance and phylogenetic analysis. The sequence of Euphlyctis hexadactylus (AY014386) was used as an out-group in the Bayesian analysis (BA). The dataset was aligned using ClustalX (Thompson et al. Citation1997) and Kimura 2 parameter (K2P) genetic distances were calculated using MEGAX program (Kumar et al. Citation2018). The BA topology was constructed in Mr. Bayes 3.1.2 (Ronquist and Huelsenbeck Citation2003) by selecting nst = 6 and rates = invgamma for GTR + G + I model estimated through Mr.Modeltest v2 (Nylander Citation2004). The phylogeny was illustrated in a web-based iTOL tool (https://itol.embl.de/) (Letunic and Bork Citation2007).
Results and discussion
Northeast India has gained worldwide attention due to the pristine natural condition, zoogeography, climates, and diverse forest cover. This region is a part of two global biodiversity hotspots with remarkable faunal diversity with endemism (Tordoff et al. Citation2011). However, apart from the checklist and taxonomic information, limited attempts were aimed to deal with molecular data to assess the herpetofaunal diversity in this region. The collected specimen was preliminarily identified as H. litoralis based on the morphological characters. The previous study reported that the mean snouts vent length (SVL) of H. litoralis is less than H. tigerinus (89.96 ± 5.92 mm in male and 101.42 ± 12.01 mm in female). The SVL of the collected female specimen was 94 mm which is within the range of H. litoralis and different from the female of H. tigerinus (113.67 ± 15.43 mm). The diagnostic feature for H. litoralis was observed in the collected individual. Two continuous distinct bands, first from anterior corner of eye through the nostrils to anterior edge of upper jaw and second along the lateral margin of upper jaw were observed in the studied specimen. Further, a distinct black margin in the inner side of the upper arm, inner metatarsal tubercle is black, and inter-orbital distance is narrower than eyelid width and inter-nostril distance were also noticed in the collected specimen. Due to the lack of bioacoustics information and avoid to wait for the upcoming seasons, we adopted the support of molecular tool to confirm the species identity. Besides, the species was described from the coastal belt of Bangladesh of about 3 m ASL and also reported from India of about 80 m ASL (Kamrup district Assam). This is the first record of the species from Mizoram state as well as from the Dampa Tiger Reserve that links from eastern India (West Bengal) to Myanmar through Bangladesh and northeast India. We provide the range extension (ca.255 km towards northeast from the type locality) and new altitude record (268 m) of H. litoralis from India.
The high-altitude adaptation allows an excellent phenomenon for examining how the animals cope with environmental stressors and accumulating genetic alterations and other physiological systems (Hutchison et al. Citation1976; Weber et al. Citation2002). The frogs are evident to acquire different genetic mechanisms correlated with the elevation distribution and high-altitude adaptation compared to other endotherms (Chen et al. Citation2013; Yang et al. Citation2016; Chen et al. Citation2018). Due to the high-altitude adaptation the physiological traits (body size and body mass), behavior such as vocal response are also switched among anurans and often hypothesized with different ecogeographical rules (Hu et al. Citation2011; Meenderink et al. Citation2017; von May et al. Citation2017). The range expansion of H. litoralis from coastal area of Bangladesh to the erratic environmental conditions at high-altitude in northeast India manifested strong and opposing selection pressures on their different life stages and influence fitness at the same time. Our results furnished further scope for additional empirical data to recognize the physiology and gene modification of this species.
The combined approach of both morphology and molecular data also evidenced that, the amphibians are one of the unique biotic components which can provide new insights on the phylogeny and diversification hypothesis (Biju and Bossuyt Citation2003; Bossuyt et al. Citation2006; Kamei et al. Citation2012). However, the knowledge of amphibian species and their genetic diversity is still under progress in northeast India. The study provides the first molecular data (470 bp mtCytb gene) of H. litoralis from India. The generated sequences showed 98.51% similarity in nucleotide BLAST search with the published database sequences generated from the type locality in Bangladesh (Hasan et al. Citation2012). The overall mean K2P genetic distance of five Hoplobatrachus species was 19% in the studied dataset. The intra-species genetic distance was ranging from 0.2% (H. tigerinus) to 4.9% (H. crassus). The inter-species genetic distance was ranging from 13.9% (H. litoralis and H. tigerinus) to 28.8% (H. crassus and Hoplobatrachus occipitalis). The Sub-Saharan African species, H. occipitalis revealed 23.6% to 28.8% genetic distance with other four species distributed in southern and southeastern Asia. All the studied Hoplobatrachus species showed monophyletic clustering in the BA phylogeny with high posterior probability support (). The targeted species, H. litoralis depicted sister relationship with H. tigerinus, as suggested in the previous studies (Hasan et al. Citation2012; Sultana et al. Citation2017; Mulcahy et al. Citation2018). The generated sequence of H. litoralis from northeast India showed 1.5% genetic distance from the generated sequences from Bangladesh. H. litoralis might have acquired these genetic changes due to the alteration of the ecological niche, which can be further demonstrated through further studies. By observing the genetic variability of H. litoralis within its range distribution, the present study encouraged extensive sampling of this species from India, Bangladesh and Myanmar to elucidate the phylogeography and gene flow.
Acknowledgements
We are thankful to the Director of Zoological Survey of India (ZSI), Ministry of Environment, Forest and Climate Change (MoEF&CC), Govt. of India for providing necessary facilities and support for the study. The first author (SK) acknowledges the fellowship grant received from the Council of Scientific and Industrial Research (CSIR) Senior Research Associateship (Scientists’ Pool Scheme) Pool No. 9072-A. The second author (HTL) would like to express his sincere thanks to the Chief Wildlife Warden, Department of Environment, Forest and Climate Change, Govt. of Mizoram for the collection permission, Ht. Decemson for helping in specimen collection.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank database at (https://www.ncbi.nlm.nih.gov) with the accession number (MT474118) which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Additional information
Funding
References
- Bickford D, Lohman DJ, Sodhi NS, Ng PK, Meier R, Winker K, Ingram KK, Das I. 2007. Cryptic species as a window on diversity and conservation. Trends Ecol Evol (Amst). 22(3):148–155.
- Biju SD, Bossuyt F. 2003. New frog family from India reveals an ancient biogeographical link with the Seychelles. Nature. 425(6959):711–714.
- Biju SD, Bossuyt F. 2009. Systematics and phylogeny of Philautus. Gistel, 1848 (Anura, Rhacophoridae) in the Western Ghats of India, with descriptions of 12 new species. Zool J Linnean Soc. 155(2):374–444.
- Biju SD, Garg S, Gururaja KV, Yogesh S, Sandeep WA. 2014. DNA barcoding reveals unprecedented diversity in Dancing Frogs of India (Micrixalidae, Micrixalus): a taxonomic revision with description of 14 new species. Ceylon J Sci (Bio Sci). 43:1–87.
- Bohra S, Bhattacharjee R, Khan N, Purkayastha J. 2019. Geographic distribution: Hoplobatrachus litoralis. Herpetol Rev. 50:521.
- Bossuyt F, Brown RM, Hillis DM, Cannatella DC, Milinkovitch MC. 2006. Phylogeny and biogeography of a cosmopolitan frog radiation: late cretaceous diversification resulted in continent-scale endemism in the family ranidae. Syst Biol. 55(4):579–594.
- Chambers EA, Hebert P. 2016. Assessing DNA barcodes for species identification in North American reptiles and amphibians in natural history collections. PLoS One. 11(4):e0154363.
- Chen W, Peng L, Jiang L, Pike DA, Friesen CR, Brown G. 2018. High altitude frogs (Rana kukonoris) adopt a diversified bet-hedging strategy in the face of environmental unpredictability. Asian Herpetol Res. 9:43–49.
- Chen W, Tang ZH, Fan XG, Wang Y, Pike DA. 2013. Maternal investment increases with altitude in a frog on the Tibetan Plateau. J Evol Biol. 26(12):2710–2715.
- Frost DR 2020. Amphibian species of the world: an online reference. Version 6.0. New York: American Museum of Natural History. [accessed 2020 May 16]. http://research.amnh.org/herpetology/amphibia/index.html
- Garg S, Biju SD. 2019. New microhylid frog genus from Peninsular India with Southeast Asian affinity suggests multiple Cenozoic biotic exchanges between India and Eurasia. Sci Rep. 9(1):1906
- Hasan MK, Kuramoto M, Islam MM, Alam MS, Khan MMR, Sumida M. 2012. A new species of genus Hoplobatrachus (Anura, Dicroglossidae) from the coastal belt of Bangladesh. Zootaxa. 3312(1):45–48.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc Biol Sci. 270(1512):313–322.
- Hu J, Xie F, Li C, Jiang J. 2011. Elevational patterns of species richness, range and body size for spiny frogs. PLoS One. 6(5):e19817
- Hutchison VH, Haines HB, Engbretson G. 1976. Aquatic life at high altitude: respiratory adaptations in the Lake Titicaca frog. Telmatobius Coleus. Respir Physiol. 27(1):115–129.
- IUCN. 2020. The IUCN Red List of Threatened Species. Version 2020.1. [accessed 2020 May 16]. https://www.iucnredlist.org
- Kamei RG, San Mauro D, Gower DJ, Van Bocxlaer I, Sherratt E, Thomas A, Babu S, Bossuyt F, Wilkinson M, Biju SD. 2012. Discovery of a new family of amphibians from northeast India with ancient links to Africa. Proc Biol Sci. 279(1737):2396–2401.
- Köhler J, Jansen M, Rodríguez A, Kok PJR, Toledo LF, Emmrich M, Glaw F, Haddad CFB, Rödel M, Vences M. 2017. The use of bioacoustics in anuran taxonomy: theory, terminology, methods and recommendations for best practice. Zootaxa. 4251(1):1–124.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kundu S, Kumar V, Tyagi K, Chandra K. 2020. The complete mitochondrial genome of the endangered Assam Roofed Turtle, Pangshura sylhetensis (Testudines: Geoemydidae): genomic features and phylogeny. PLoS One. 15(4):e0225233.
- Letunic I, Bork P. 2007. Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23(1):127–128.
- Mahony S, Foley NM, Biju SD, Teeling EC. 2017. Evolutionary history of the Asian horned frogs (Megophryinae): integrative approaches to timetree dating in the absence of a fossil record. Mol Biol Evol. 34(3):744–771.
- Meenderink SWF, Quiñones PM, Narins PM, 2017. Behavioural tuning in a tropical amphibian along an altitudinal gradient. Biol Lett. 13(12):20170317. 1.
- Mondal K, Purkayastha J, Chaudhuri A. 2018. Geographic distribution: Hoplobatrachus litoralis. Herpetol Rev. 49:500.
- Mulcahy DG, Lee JL, Miller AH, Chand M, Thura MK, Zug GR. 2018. Filling the BINs of life: report of an amphibian and reptile survey of the Tanintharyi (Tenasserim) Region of Myanmar, with DNA barcode data. ZK. 757:85–152.
- Nylander J. 2004. Mr. Modeltest v2, program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University.
- Ortega-Andrade HM, Rojas-Soto OR, Valencia JH, Espinosa de los Monteros A, Morrone JJ, Ron SR, Cannatella DC. 2015. Insights from integrative systematics reveal cryptic diversity in Pristimantis frogs (Anura: Craugastoridae) from the Upper Amazon Basin. PLoS One. 10(11):e0143392.
- Padial JM, Miralles A, De la Riva I, Vences M. 2010. The integrative future of taxonomy. Front Zool. 7:16.
- Purkayastha J, Basak S. 2018. Hoplobatrachus litoralis (Anura: Dicroglossidae) in India. Hamadryad. 38:25–26.
- Roelants K, Gower DJ, Wilkinson M, Loader SP, Biju SD, Guillaume K, Moriau L, Bossuyt F. 2007. Global patterns of diversification in the history of modern amphibians. Proc Natl Acad Sci USA. 104(3):887–892.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Sultana N, Igawa T, Islam MM, Hasan M, Alam MS, Komaki S, Kawamura K, Khan MM, Sumida M. 2017. Inter- and intra-specific genetic divergence of Asian tiger frogs (genus Hoplobatrachus), with special reference to the population structure of H. tigerinus in Bangladesh. Genes Genet Syst. 91(4):217–227.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25(24):4876–4882.
- Tordoff AW, Bezuijen MR, Duckworth JW, Fellowes JR, Koenig K, Pollard EHB, Royo AG. 2011. Ecosystem profile: Indo-Burma biodiversity hotspot, 2011 update. Washington (DC): Critical Ecosystem Partnership Fund. p. 381.
- Tyagi K, Kumar V, Singha D, Chandra K, Laskar BA, Kundu S, Chakraborty R, Chatterjee S. 2017. DNA barcoding studies on thrips in India: cryptic species and species complexes. Sci Rep. 7(1):4898
- Van Bocxlaer I, Biju SD, Willaert B, Giri VB, Shouche YS, Bossuyt F. 2012. Mountain-associated clade endemism in an ancient frog family (Nyctibatrachidae) on the Indian subcontinent. Mol Phylogenet Evol. 62(3):839–847.
- Van Bocxlaer I, Roelants K, Biju SD, Nagaraju J, Bossuyt F. 2006. Late Cretaceous vicariance in Gondwanan amphibians. PLoS One. 1:e74
- Verma SK, Singh L. 2002. Novel universal primers establish identity of an enormous number of animal species for forensic application. Mol Ecol Notes. 3(1):28–31.
- von May R, Catenazzi A, Corl A, Santa-Cruz R, Carnaval AC, Moritz C. 2017. Divergence of thermal physiological traits in terrestrial breeding frogs along a tropical elevational gradient. Ecol Evol. 7(9):3257–3267.
- Weber RE, Ostojic H, Fago A, Dewilde S, Hauwaert MV, Moens L, Monge C. 2002. Novel mechanism for high-altitude adaptation in hemoglobin of the Andean frog Telmatobius peruvianus. Am J Physiol Regul Integr Comp Physiol. 283(5):R1052–R1060.
- Yang W, Qi Y, Fu J. 2016. Genetic signals of high-altitude adaptation in amphibians: a comparative transcriptome analysis. BMC Genet. 17(1):134.
