Abstract
In this study, the complete chloroplast genome of Tradescantia pallida (Rose) D.R.Hunt was investigated. The whole chloroplast genome sequence is 166,086 bp in length, which consists of a 94,376 bp large single copy (LSC) and an 18,678 bp small single copy (SSC) regions, separated by a pair of 26,518 bp inverted repeat (IR) regions. The chloroplast genome of T. pallida encodes 131 annotated known unique genes including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis based on chloroplast genome sequences demonstrated that T. pallida is most closely related to Belosynapsis ciliate.
Keywords:
Tradescantia pallida (Rose) D.R.Hunt is a material for plant morphology and physiology study, which mainly distribute in tropical countries. The staminal hairs of T. pallida played an important role in studying cell-to-cell passage of small molecules (Tucker and Boss Citation1996). The flower of T. pallida can be used to detect radiation. In addition, T. pallida is also a good material for observing thick-angled tissues, thin-walled tissues, threaded vessel, cytoplasmic flow and posterior inclusion crystal (Crispim et al. Citation2012). In addition, the whole herb of T. pallida can be used as traditional medicines, which can activate blood, resolve stasis and remove toxin for detumescence (Dash et al. Citation2020), and has a significant effect on the treatment of snake bites, bruises, swelling, and rheumatism. However, there are few reports about the complete chloroplast genome of T. pallida.
The fresh leaves of T. pallida were collected from Xianyang (Shaanxi, China; 34°19’N, 108°44’E). Voucher specimen (610402191202001LY) was deposited in the herbarium of traditional Chinese Medicine, Shaanxi University of Chinese Medicine. Fresh leaves were silica-dried and taken to the laboratory. Whole genome sequencing was performed using Illumina Hiseq X10 platform (Illumina, San Diego, CA). Sequencing platform based on Sequencing By Synthesis (SBS) technology. A total of 47,670,410 clean reads were produced. The whole chloroplast genome was assembled using SPAdes v3.11.1 (http://cab.spbu.ru/software/spades/) (Bankevich et al. Citation2012) and then annotated by CpGAVAS and DOGMA (http://dogma.ccbb.utexas.edu/) (Liu et al. Citation2012). A physical map of the chloroplast genome was generated by OGDRAW.
The complete chloroplast genome of T. pallida (GenBank accession number: MT527961) is 166,086 bp in length, containing a large single-copy region (LSC) of 94,376 bp, a small single-copy region (SSC) of 18,678 bp, and two inverted repeat (IR) regions of 26,518 bp, and the GC content is 35.15%. It contains 86 protein-coding genes, 37 tRNA genes and 8 rRNA genes. Among which 19 genes duplicated in the IR regions, including seven protein-coding genes (rps7, rps12, rps19, rpl2, rpl23, ndhB, and ycf2), eight tRNAs (trnA-UGC, trnH-GUG, trnl-GAU, trnl-CAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and four rRNAs (4.5S, 5S, 16S, and 23S rRNA). In addition, 15 genes contain one intron, two genes (ycf3 and clpP) exhibit two introns.
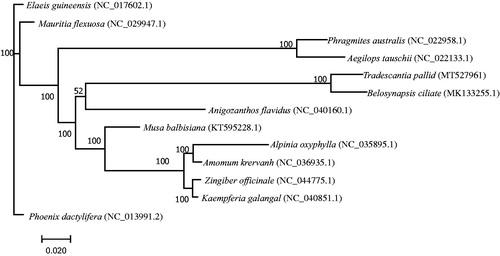
Genome sequences were aligned using MAFFT (Yamada et al. Citation2016), and a maximum-likelihood phylogenetic tree including T. pallida and other 12 reported species (Cui and Liang Citation2019; Guo et al. Citation2019) was constructed using RAxML Version 8 with 1000 bootstrap replicates (Stamatakis Citation2014). The result of phylogeny analysis indicated that T. pallida is most closely related to Belosynapsis ciliate from the same family Commelinaceae (). This complete chloroplast genome sequence of T. pallida will provide useful information for future studies in solving the phylogenetic relationships of Commelina species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, accession number MT527961.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Crispim BA, Vaini JO, Grisolia AB, Teixeira TZ, Mussury RM, Seno LO. 2012. Biomonitoring the genotoxic effects of pollutants on Tradescantia pallida (Rose) D.R. Hunt in Dourados, Brazil. Environ Sci Pollut Res. 19(3):718–723.
- Cui Y, Liang RF. 2019. The chloroplast genome sequence of Commelina communis (Commelinaceae). Mitochondrial DNA Part B. 4(2):2631–2632.
- Dash GK, Hassan N, Hashim MHB, Muthukumarasamy R. 2020. Pharmacognostic study of Tradescantia pallida (Rose) DR Hunt leaves. Res J Pharm Technol. 13(1):233–236.
- Guo W, Xu YF, Wu W, Ye YH, Chen P. 2019. The complete chloroplast genome sequence of Belosynapsis ciliata (Blume) R. S. Rao (Commelinaceae). Mitochondrial DNA Part B. 4(2):2373–2374.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Liu XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13(1):715.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tucker EB, Boss WF. 1996. Mastoparan-induced intracellular Ca2+ fluxes may regulate cell-to-cell communication in plants. Plant Physiol. 111(2):459–467.
- Yamada KD, Tomii K, Katoh K. 2016. Application of the MAFFT sequence alignment program to large data-reexamination of the usefulness of chained guide trees. Bioinformatics. 32(21):3246–3251.