Abstract
In this study, the mitochondrial genome (mitogenome) of Holothuria fuscocinerea was unraveled to be 15,890 bp in length, containing 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes. The PCGs were initiated by four initiation codons (ATG, TAC, ATC, and ATA). Only one PCG (nad6) and five tRNA genes (tRNASer(UCN), tRNAGln, tRNAAla, tRNAVal, and tRNAAsp) were encoded on the light chain, and the other genes were encoded on the heavy chain. A phylogenetic tree constructed with 16S rRNA sequences showed that H. fuscocinerea is most closely related to H. leucospilota.
Holothuria fuscocinerea (Echinodermata: Holothuroidea, H. fuscocinerea) is naturally distributed near boulders, corals, and seaweed clumps in the West Pacific, East Africa, Australia, and Southeastern China (Liao Citation1997), and it might play important roles in maintaining a healthy coral reef ecosystem (Birkeland Citation1989; Schneider et al. Citation2011).
Compared with whole mitochondrial genes, a fragment of a single mitochondrial gene such as 16S rRNA can provide more information in terms of identification and evolution (Kerr et al. Citation2005; Byrne et al. Citation2010; Liu et al. Citation2016; Zou et al. Citation2017; Liu et al. Citation2018). The classification of sea cucumber from the mitochondrial genome (mitogenome) level would be more accurate than the aspect of phenotypic characteristics such as the morphology of the tentacles, the endoskeleton, and the calcareous ring (Liao Citation1997; Rowe and Richmond Citation2004).
Holothuria fuscocinerea was obtained from Daya Bay (N22°35′, E114°31′), Shenzhen, Guangdong, China. The specimen was stored in the Marine Biotechnology and Disease Control Laboratory of the South China Sea at the Chinese Academy of Sciences in Guangzhou, China (MBDC170711124). Total DNA was extracted and sent to BGI Genomics Co., Ltd., China, for sequencing. After aligning, splicing, correction, and identification of protein-coding genes and tRNA genes, the obtained mitogenome DNA sequence was analyzed by phylogenetic analysis.
The mitogenome of H. fuscocinerea (MK391177) show a double-strand molecule of 15,890 bp (33.1% A, 27.3% T, 23.9% C, and 15.7% G), including a set of 22 tRNA genes that varied from 61 bp (tRNALys) to 72 bp (tRNALeu(UUR)) in length (the total length of tRNAs was 1,511 bp), 13 protein-coding genes that consisted of 3,690 codons, 2 rRNAs (lrRNA: GC% = 38.07%, srRNA: GC% = 43.48%), and a putative noncoding control region between tRNAThr and tRNAPro (GC% = 59.01%).
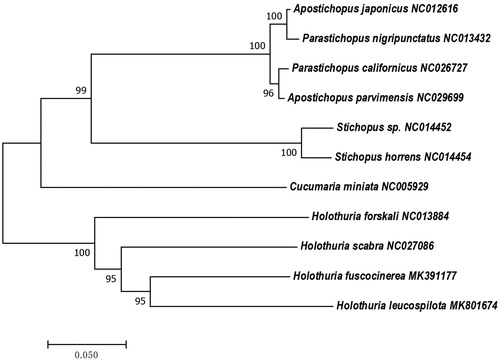
The phylogenetic analyzing demonstrated that H. fuscocinerea most closely related to H. leucospilota (). The phylogenetic tree exhibits consistent topology, indicating that the interspecific relationships among holothuroids are monophyletic (). Identifying the taxonomy of organisms from the perspective of mitochondrial genes avoids convergent evolution, which means that distant species can evolve similar phenotypes.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference number MK391177.
Additional information
Funding
References
- Birkeland C. 1989. The influence of echinoderms on coral-reef communities. Echinoderm Studies. 3:1–79.
- Byrne M, Rowe F, Uthicke S. 2010. Molecular taxonomy, phylogeny and evolution in the family Stichopodidae (Aspidochirotida: Holothuroidea) based on COI and 16S mitochondrial DNA. Mol Phylogenet Evol. 56(3):1068–1081.
- Kerr AM, Janies D, Clouse R, Samyn Y, Kuszak J, Kim J. 2005. Molecular phylogeny of coral-reef sea cucumbers (Holothuriidae: Aspidochirotida) based on 16S mitochondrial ribosomal DNA sequence. Mar Biotechnol. 7(1):53–60.
- Liao Y. 1997. Fauna sincia: Phylum Echinodermata Class Holothuroidea. Beijing: Science Press.
- Liu N, Li N, Yang P, Sun C, Fang J, Wang S. 2018. The complete mitochondrial genome of Damora sagana and phylogenetic analyses of the family nymphalidae. Genes Genomics. 40(1):109–122.
- Liu QN, Chai XY, Bian DD, Ge BM, Zhou CL, Tang BP. 2016. The complete mitochondrial genome of fall armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae). Genes Genom. 38(2):205–216.
- Rowe FWE, Richmond MD. 2004. A preliminary account of the shallow-water echinoderms of Rodrigues, Mauritius, western Indian Ocean. J of Natural Hist. 38(23):3273–3314.
- Schneider K, Silverman J, Woolsey E, Eriksson H, Byrne M, Caldeira K. 2011. Potential influence of sea cucumbers on coral reef CaCO3 budget: a case study at One Tree Reef. J Geophys Res. 116(G4):1–6.
- Zou Y-C, Xie B-W, Qin C-J, Wang Y-M, Yuan D-Y, Li R, Wen Z-Y. 2017. The complete mitochondrial genome of a threatened loach (Sinibotia reevesae) and its phylogeny. Genes Genom. 39(7):767–778.