Abstract
In this study, we sequenced the mitogenome of Habrobracon hebetor, and obtained almost complete mitogenome of it. The mitogenome contains 14,629 bp which consists of 13 protein-coding genes (PCGs), 20 transfer RNA genes (trnI and trnM are absent), and 2 ribosomal RNA genes (GenBank accession no. MT558946). Gene rearrangement events occurred in this species, five tRNA genes with changes in positions or/and directions are found. All of 13 PCGs started with ATN. Eight PCGs used the typical stop codon ‘TAA’, five PCGs terminated with incomplete stop codons (T). Phylogenetic analyses within the Cyclostomes were performed based on mitochondrial PCGs.
Habrobracon hebetor (Say) (Hymenoptera: Braconidae: Braconinae: Braconini) is a cosmopolitan gregarious, idiobiont, larval ectoparasitoid of pyralid and noctuid moths (Magro and Parra Citation2001; Abedi et al. Citation2012). It is considered one of the most important biological control agents of pyralid moths in stored products. To date, no mitogenome had been studied for the tribe, and no complete mitogenome of Braconinae reported.
In this study, adult samples of H. hebetor were obtained in the insectarium of Environment and Plant Protection Institute, China Academy of Tropical Agriculture Sciences, Hainan, China (110°20′9″N, 19°59′21″E). The specimens were deposited at −20 °C in the herbarium of Post-Entry Quarantine Station for Tropical Plant, Haikou Customs District, PR China (speciemen accesion number IN07070201-0000-0010). A single individual was used for DNA extraction.
The mitogenome sequence of H. hebetor only obtained 14,629 bp (GenBank accession number MT558946), contained 13 protein-coding genes (PCGs), 20 tRNA genes, and 2 rRNA genes. Two tRNA (trnI and trnM) and A + T rich region were absent. The nucleotide composition of the known mitogenome sequence was biased toward AT 84.8%, and the base composition was 42% A, 42.8% T, 6.8% G, and 8.4% C.
Gene rearrangement events occurred in this species. All rearranged genes were tRNA. The tRNA arrangement patterns of trnW–trnY–trnC and trnD–trnH(–)–trnK in the sequenced region were found. These rearrangement patterns of tRNAs were consistent with previous results (Dowton Citation1999; Wei et al. Citation2010; Zhang et al. Citation2020).
The length of 13 PCGs was 10,877 bp, all PCGs started with ATN, ATA for nad2, cox1, cox2, atp8, cox3, nad3, and nad5; ATG for atp6, nad4, nad6, and cob; ATT for nad4l and nad1. Five PCGs (atp6, nad3, nad5, nad4, and cytb) terminated with incomplete stop codons (T), the rest PCGs used the typical stop codon ‘TAA’. All of the 20 tRNAs have the usual clover-leaf secondary structure, except for trnS1. All tRNAs had normal lengths, which varied from 64 to 71 bp. The 16S rRNA was 1291 bp long with an AT content of 88.5%, while the 12S rRNA only sequenced 365 bp long.
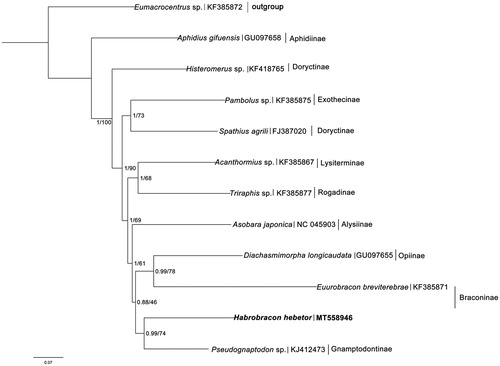
We performed phylogenetic analyses of H. hebetor with 10 other species representing 10 subfamilies of Cyclostomes and one species from Helconinae as outgroup based on only 12 PCGs on account of most known mitogenomes of Cyclostomes lost the gene ND2 (). The analyses were performed with Bayesian inference MrBayes 3.2.3 (Ronquist et al. Citation2012) and maximum likelihood in RAxML 8.2.10 (Stamatakis Citation2014). The results of phylogenetic analysis within Cyclostomes were similar to the previous study (Li et al. Citation2016). However, as the only two mitochondrial genomes of Braconinae reported so far, H. hebetor and Euurobracon breviterebrae did not form a monophyletic lineage in the phylogenetic tree, and the result showed they were close to Opiinae and Gnamptodontinae. These results may indicate that more mt-genome sequences are required to resolve the phylogenic relationships within Cyclostomes in further studies.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number [MT558946], or available from the corresponding author.
Additional information
Funding
References
- Abedi Z, Saber M, Gharekhani G, Mehrvar A, Mahdavi V. 2012. Effects of azadirachtin, cypermethrin, methoxyfenozide and pyridalil on functional response of Habrobracon hebetor Say (Hym.: Braconidae). J Plant Prot Res. 52(3):353–358.
- Dowton M. 1999. Relationships among the cyclostome braconid (Hymenoptera: Braconidae) subfamilies inferred from a mitochondrial tRNA gene rearrangement. Mol Phylogenet Evol. 11(2):283–287.
- Li Q, Wei SJ, Tang P, Wu Q, Shi M, Sharkey MJ, Chen XX. 2016. Multiple lines of evidence from mitochondrial genomes resolve phylogenetic relationships of parasitic wasps in Braconidae. Genome Biol Evol. 8(9):2651–2662.
- Magro SR, Parra JRP. 2001. Biology of ectoparasitoid Bracon hebetor Say, 1857 (Hymenoptera: Braconidae) on seven Lepidopteran species. Sci Agric (Piracicaba, Braz). 58(4):693–698.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wei SJ, Shi M, Sharkey MJ, van Achterberg C, Chen XX. 2010. Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to Holometabolous insects. BMC Genomics. 11:371.
- Zhang X, Li CQ, Pan ZQ, Zhu JC, Wang ZZ, Shi M, Chen XX, Huang JH. 2020. The complete mitochondrial genome of Asobara japonica (Hymenoptera: Braconidae). Mitochondrial DNA B. 5(2):1279–1281.