Abstract
The complete mitochondrial genome (mitogenome) of Quasipaa exilispinosa (Anura: Dicroglossidae) we sequenced can provide critical information for mitogenome evolution. This mitogenome with 17,046 bp long contains 13 protein-coding genes (PCGs), 22 transfer RNA genes, two ribosomal RNA genes, and one non-coding regions. The overall AT content of 58.6%. The data using Bayesian phylogenetic inference supports the close relationship with Q. spinosa from the genus Quasipaa. Our results will be helpful for detail study on phylogenetic relationships of the related taxa.
Lesser spiny frog (Quasipaa exilispinosa) is a medium-sized frog, which name comes from their skin rough with tubercules and granules. They are mainly distributed in mountain stream in a wide range of altitudes between 500 and 1400 m (Liang et al. Citation2012). In this study, we sequenced the mitochondrial genome of Q. exilispinosa (GenBank accession No. MT561179), and found that it had a close relationship with the other species from the genus Quasipaa.
The specimen of Q. exilispinosa was collected in Mount Wuyi National Park from north of Fujian Province in China (117°44′21″E, 27°35′46″N) and preserved with 95% ethanol at temperature −20 °C in the Lishui University (Specimen accession number: LSU20200418001WY). The genomic DNA extracted from muscular tissue using EasyPure@ Genomic DNA Kit according to the manufacturer’s instructions (TransGen Biotech Co, Beijing, China), and Illumina sequencing was done by Novogene Bioinformatics Technology Co. Ltd. (Tianjin, China). The high-throughput sequence data obtained were then assembled using the SeqMan (DNAstar, Inc) to generate the complete mitochondrial genome, and annotated using MITOS Web Server (Bernt et al. Citation2013). Raw sequence data (18.30 Gb) were deposited into Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/) with the accession no. SRR11921102, and clean data without sequencing adapters were de novo assembled using the NOVOPlasty 3.7 (Dierckxsens et al. Citation2017).
The complete mitogenome of Q. exilispinosa was a circular molecule with 17,046 bp long, encoded 13 protein-coding genes (PCGs), two ribosomal RNAs genes (rrnL and rrnS), 22 transfer RNAs genes, and a non-coding control region with 1559 bp. The overall nucleotide composition was 29.3% A, 27.2% C, 14.2% G, and 29.4% T. The ATP8 was the shortest gene with 162 bp among the 13 PCGs, while the ND5 was the longest with 1824 bp. Nine genes (ND1, COII, ATP8, ATP6, COIII, ND4L, ND4, ND5, and Cytb) began with codon ATG, two genes (ND3, ND6) began with ATA, and two genes (ND2, COI) began with ATT and GTG, respectively. Seven genes (ND2, COI, ATP8, ND4L, ND5, ND6, and Cytb) end with complete stop codons (TAG, AGG, and TAA), and the other six genes have incomplete stop codons with TA or T––. Anderson et al. (Citation1981) conferred that they could be form TAA by post-transcription polyadenylation.
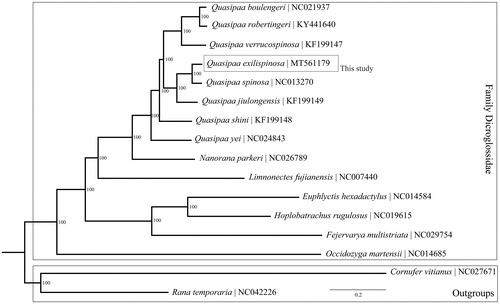
We have constructed phylogenetic tree based on 13 PCGs of 14 species belong to the homologous Family Dicroglossidae and two other species (Rana temporaria and Cornufer vitianus) as the outgroups (). The Bayesian Inference (BI) tree was constructed on MrBayes version 3.2.2 (Ronquist et al. Citation2012) and the best-fit models of molecular evolution were chosen under the Akaike Information Criterion in MrModeltest version 2.3 (Nylander Citation2004). Our result shows that Q. exilispinosa was positioned near Q. Spinosa, which means they may derive from a common ancestor.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in [NCBI] at www.ncbi.nlm.nih.gov, reference number (MT561179).
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290(5806):457–465.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Liang F, Chang-Yuan Y, Jian-Ping J. 2012. Colored atlas of Chinese amphibians and their distributions. China: Sichuan Publishing Group. p. 465. (In Chinese)
- Nylander JA. 2004. MrModeltest version 2. Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala University.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.