Abstract
We reported the partial mitochondrial genome (mitogenome) for Hyla sanchiangensis (Anura: Hylidae), a arboreal frog and endemic in China. The length of partial mitogenome of H. sanchiangensis was 15,664 bp, and contained PCGs (COX1-3, ND1-6, ND4L, ATP6, ATP8 and CYTB), 2 ribosomal RNA genes, 22 transfer RNA genes, and 2 non-coding regions of a L-strand replication origin and a partial loop region. The overall base composition of the sequence is 29.91% A, 29.86% T, 14.58% G, and 25.65% C, with a total A + T content of 59.77%. The result of phylogenetic analysis showed that H. sanchiangensis formed a clade with other species belonging to the genus of Hyla. This mitogenome data could help in evolutionary biology and population genetics of the Hylid species.
There are currently 17 species of Hyla (Anura: Hylidae) distributed in Eurasia (Frost Citation2020). Eight species of Hyla are currently known in China, and six of which are endemic to China (AmphibiaChina Citation2020). The Sanchiang Tree Toad (Hyla sanchiangensis) is a endemic Chinese arboreal frog, which is distributed in southeastern of China (AmphibiaChina Citation2020). At GenBank, only three mitochondrial genomes (mitogenomes) are available for Hyla speices. Here, we attempted to sequence the complete mitogenome of H. sanchiangensis via next-generation sequencing, and retrieved successfully the genetic information about partial mitogenome of the above mentioned species.
The specimen (LSU20200425002JL) of H. sanchiangensis was collected in Jiulongshan National Nature Reserve (28.37°N, 118.90°E), Zhejiang, China, and deposited in the Museum of Laboratory of Amphibian Diversity Investigation (ADI) at Lishui University. Total DNA was extracted from muscle tissue of H. sanchiangensis using EasyPure Genomic DNA Kit (TransGen Biotech Co, Beijing, China). The mitogenome of H. sanchiangensis was acquired by Illumina NovaSeq 6000 (Novogene Bioinformatics Technology Co. Ltd., Tianjin, China) for PE 2 × 150 BP sequencing. Raw sequence data (16.25 G) was deposited in NCBI’s Sequence Read Archive (SRA; accession: SRR11921085). We used the NOVO Plasty 3.7 to de novo assembled the clean data without sequencing adapters (Dierckxsens et al. Citation2017).
This study reported a 15,664 bp long sequence of H. sanchiangensis partial mitogenome (GenBank accession: MT561180), which containing 13 PCGs (COX1-3, ND1-6, ND4L, ATP6, ATP8 and CYTB), 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and 2 non-coding regions of a L-strand replication origin and a partial loop region. The overall base composition of the sequence is 29.91% A, 29.86% T, 14.58% G, and 25.65% C, with a total A + T content of 59.77%. Similar to the mitogenomes of other Hyla species, the AT content is higher than the GC content (Zhang et al. Citation2005; Kang et al. Citation2016; Ye et al. Citation2016). The H-strand encodes12 PCGs, 14 tRNA genes, and 2 rRNA genes, while the ND6, and 8 other tRNA genes are encoded on the L-strand. Among the 13 PCGs, the ND5 was the longest (1803 bp), while the ATP8 was the shortest (165 bp). Codon usage analysis of H. sanchiangensis showed that three kinds of start codons (ATG, ATT, and TTG) and four kinds of stop codons (TAA, AGA, TAG, and T) were used. The 12S rRNA (937 bp) and 16S rRNA genes (1596 bp) are located between tRNAPhe and tRNAVal and between tRNAVal and tRNALeu, respectively. The 22 tRNA genes varied in size from 64 to 73 bp.
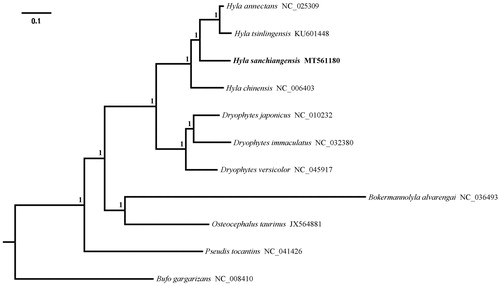
Phylogenetic analysis was inferred from available mitogenomes of H. sanchiangensis and other 9 Hylid species based on 13 PCGs with Bufo gargarizans (Anura: Bufonidae) as the outgroup using Bayesian inference (BI) methods. The optimal substitution model (GTR + I + G) was implemented via jModelTest (Darriba et al. Citation2012). Four parallel runs of Markov Chain Monte Carlo (MCMC) were analyzed for 1,000,000 generations, sampling every 1000 generations and discarded 1000 trees as burn-in. The results of phylogenetic relationship showed that H. sanchiangensis formed a clade with other species (H. annectans, H. chinesis, and H. tsinlingensis) belonging to the genus of Hyla, the genus Hyla was closely related with the genus Dryophytes than to other genera, and the genus Pseudis was a basal clade relative to others within Hylidae (). This partial mitogenome sequence of H. sanchiangensis could help in evolutionary biology, population genetics of the Hylid species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at www.ncbi.nlm.nih.gov reference number [MT561180].
Additional information
Funding
References
- AmphibiaChina 2020. The database of Chinese amphibians. Kunming (China): Kunming Institute of Zoology (CAS). Available from: http://www.amphibiachina.org/. (accessed 4 June 2020)
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Frost DR. 2020. Amphibian species of the world: an online reference, version 6.0. New York (NY): American Museum of Natural History. [accessed 2020 June 4]. http://research.amnh.org/herpetology/amphibia/index.php/
- Kang X, Sun ZL, Guo WB, Wu J, Qian LF, Pan T, Wang H, Li K, Zhang BW. 2016. Sequencing of complete mitochondrial genome for Tsinling Tree Toad (Hyla tsinlingensis). Mitochondrial DNA B. 1(1):466–467.
- Ye LT, Zhu CC, Yu DN, Zhang YP, Zhang JY. 2016. The complete mitochondrial genome of Hyla annectans (Anura: Hylidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(3):1593–1594.
- Zhang P, Zhou H, Chen YQ, Liu YF, Qu LH. 2005. Mitogenomic perspectives on the origin and phylogeny of living amphibians. Syst Biol. 54(3):391–400.