Abstract
Aconitum flavum, a traditional Chinese medicine. The complete chloroplast genome sequence is 155,654 bp in length, with one large single copy region of 86,390 bp, one small single copy region of 16,968 bp, and two inverted repeat (IR) regions of 26,148 bp. It contains 129 genes, including 83 protein-coding genes, 8 ribosomal RNA, and 37 transfer RNA. Phylogenetic tree shows that this species is a sister to A. brachypodum.
The Aconitum consisting of over 200 species distributed in China, Most of them grow among high altitudes. Such as A. kusnezoffii, A. carmichaelithe and A. flavum (Xiao Citation2006). In traditional Chinese medicine, Aconitum have similar pharmacological actions and are commonly applied for various inflammatory diseases, such as rheumatic fever, painful joints, gastroenteritis, diarrhea, edema, bronchial asthma and various tumors (Singhuber et al. Citation2009; Qin et al. Citation2012). However, due to anthropogenic over exploitation and decreasing distributions, this species needs reagent conservation. Knowledge of the genetic information of this species would contribute to the formulation of protection strategy. In this study, we assembled and characterized the complete chloroplast (cp) genome sequence of A. flavum.
Fresh leaves of A. flavum were collected from Nanhuashan mountain (Zhongwei, Ningxia, China; coordinates: 105°36′E, 36°26′N) and dried with silica gel. The voucher specimen was stored in Sichuan University Herbarium with the accstion number of QTPLJQ13383066. Total genomic DNA was extracted with a modified CTAB method (Doyle and Doyle Citation1987). First, we obtained 10 million high quality pair-end reads for A. flavum, and after removing the adapters, the remained reads were used to assemble the complete chloroplast genome by NOVOPlasty (Dierckxsens et al. Citation2017). The complete chloroplasts genome sequence of A. brachypodum was used as a reference. Plann v1.1 (Huang and Cronk Citation2015) and Geneious v11.0.3 (Kearse et al. Citation2012) were used to annotate the chloroplasts genome and correct the annotation.
The total plastome length of A. flavum (MT571464) is 155,654 bp, exhibits a typical quadripartite structural organization, consisting of a large single copy (LSC) region of 86,390 bp, two inverted repeat (IR) regions of 26,148 bp and a small single copy (SSC) region of 16,968 bp. The cp genome contains 129 complete genes, including 83 protein-coding genes (83 PCGs), 8 ribosomal RNA genes (4 rRNAs), and 37 tRNA genes (37 tRNAs). Most genes occur in a single copy, while 13 genes occur in double, including 7 tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and 6 PCGs (rps7, rpl2, rpl23, ndhB, ycf2, ycf15). The overall GC content of cp DNA is 38.1%, the corresponding values of the LSC, SSC, and IR regions are 36.2, 32.6, and 43.0%.
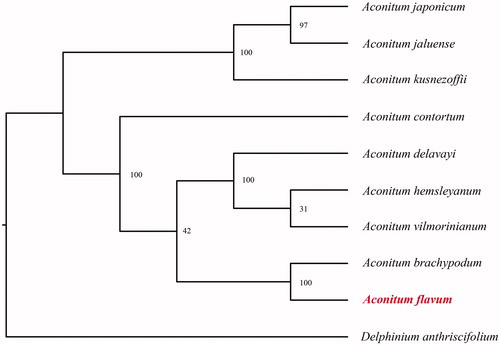
In order to further clarify the phylogenetic position of A. flavum, plastome of nine representative Aconitum species were obtained from NCBI to construct the plastome phylogeny, with Delphinium anthriscifolium as an outgroup. All the sequences were aligned using MAFFT v.7.313 (Katoh and Standley Citation2013) and maximum likelihood phylogenetic analyses were conducted using RAxML v.8.2.11 (Stamatakis Citation2014). The phylogenetic tree shows that Aconitum clade were identified as two subclades. A. japonicum, A. jaluense and A. kusnezoffii together one clustered. Remian Aconitum species together another clustered, While Aconitum brachypodum clustered together with A. flavum in this subclade ().
Figure 1. Phylogenetic relationships of Aconitum species using whole chloroplast genome. GenBank accession numbers: Aconitum brachypodum (NC_041579), Aconitum contortum (NC_038098), Aconitum delavayi (NC_038097), Aconitum episcopale (NC_038096), Aconitum hemsleyanum (NC_038095), Aconitum jaluense (KT820669), Aconitum japonicum (KT820670), Aconitum kusnezoffii (MK569468), Aconitum vilmorinianum (NC_038094), Delphinium anthriscifolium (MK253461).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT571464.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for anno-tating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Qin Y, Wang JB, Zhao YL, Shan LM, Li BC, Fang F, Jin C, Xiao XH. 2012. Establishment of a bioassay for the toxicity evaluation and quality control of Aconitum herbs. J Hazard Mater. 199–200:350–357.
- Singhuber J, Zhu M, Prinz S, Kopp B. 2009. Aconitum in traditional Chinese medicine: a valuable drug or an unpredictable risk. J Ethnopharmacol. 126(1):18–30.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Xiao PG. 2006. A pharmacophylogenetic study of Aconitum L. (Ranunculaceae) from China. Acta Phytotaxonomica Sinica. 44(1):1–46.
