Abstract
The complete mitochondrial genome of the citrus leafminer, Phyllocnistis citrella (Lepidoptera: Gracillariidae) was sequenced for the first time, which was 15,416 bp in length and with an A + T content of 81.9%. The common set of 37 genes (13 PCGs, 22 tRNA genes, and two rRNA genes) were all annotated. Most PCGs had standard ATN start codons and TAN stop codons. Most tRNA genes exhibited typical cloverleaf secondary structures, whereas the dihydrouridine (DHU) arm of trnSer (AGN) was reduced. The mitochondrial phylogenetic analyses consistently recovered the relationships within Gracillariidae.
The citrus leafminer (CLM), Phyllocnistis citrella Stainton Citation1856 (Lepidoptera: Gracillariidae), is a widespread pest species of citrus and related Rutaceae in Asia. Accumulation of new molecular data of P. citrella is a vital step to find new control strategies for this pest from a genetic view. However, the mitogenome of P. citrella is still unknown, which is a barrier for us to understand the relationship between P. citrella and its related lepidopteran pests. In this study, the complete mitogenome P. citrella is determined and reported for the first time. The sequence is accessible in GenBank with the accession number MN792920. The mitogenomic structure, nucleotide compositions, codon usages of PCGs, secondary structures of tRNA genes and the control region were analyzed to better understand the genetic characters of this important pest. The mitogenomic phylogeny of Gracillariidae was also investigated based on the newly obtained data.
The male adult samples of P. citrella were collected from Nanchong City, Sichuan Province of China (30.82 N, 106.09 E) in June 2019; all specimens and DNA samples are stored in the Insect Collection of Sichuan Academy of Agricultural Sciences (ICSAAS No. ICSAAS-LEP-1), Chengdu, China. The complete mitogenome of P. citrella is a typical double-strand circular molecule with 15,416 base pairs and the standard set of 37 genes (13 PCGs, 22 tRNA genes, and two rRNA genes). The only control region of P. citrella is short with only 405 bp, but its A + T content is the highest (94.1%) in the mitogenome. The mitochondrial genes of P. citrella are rearranged, exhibiting the trnMet-trnIle-trnGln rearrangement. The whole mitogenome of P. citrella is strongly biased toward A and T nucleotides (81.9%) with negative AT-skew and GC-skew values. Most PCGs utilize the standard ATN start codon except for the special start codon CGA used by cox1. Most PCGs terminate with the complete stop codon TAA, but cox1, cox2, and cox3 end with an incomplete stop codon T. Most of the tRNA genes can fold into cloverleaf secondary structures, but the TΨC loops of trnPhe and trnTyr are reduced and the dihydrouridine (DHU) arm of trnSer is reduced into a small loop. The large ribosomal RNA (rrnL) gene is 1370 bp in length, with an A + T content of 85.8%. The small ribosomal RNA (rrnS) gene is 761-bp long, with an A + T content of 86.1%.
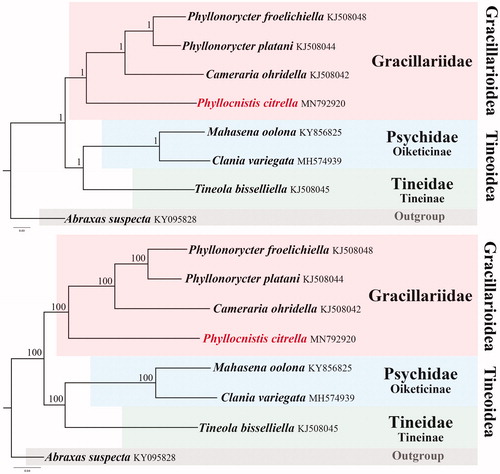
The phylogenetic position of P. citrella in Gracillariidae was investigated using BI and ML analyses, which generated identical topological structures (). In both trees, P. citrella was closely grouped with other members of Gracillariidae. The two species of Phyllonorycter were grouped in the same clade, indicating the efficiency of mitogenome data in phylogenetic reconstructions. The two families, Psychidae and Tineidae were supported as sister groups in Tineoidea. Each family studied herein was recovered as monophyletic with high support (posterior probability = 1, bootstrap value = 100). The reconstructed topology of the three families is identical with the results of Robinson (Citation1988) based on morphological characters, and consistent with the mitogenomic phylogeny of Jeong et al. (Citation2018) and Chen et al. (Citation2019).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in ‘NCBI’ at https://www.ncbi.nlm.nih.gov/, reference number MN792920.
Additional information
Funding
References
- Chen L, Liao CQ, Wang X, Tang SX. 2019. The complete mitochondrial genome of Gibbovalva kobusi (Lepidoptera: Gracillariidae). Mitochondrial DNA B. 4(2):2769–2770.
- Jeong JS, Kim MJ, Kim SS, Kim I. 2018. Complete mitochondrial genome of the female-wingless bagworm moth, Eumeta variegata Snellen, 1879 (Lepidoptera: Psychidae). Mitochondrial DNA B. 3(2):1037–1039.
- Robinson GS. 1988. A phylogeny for the Tineoidea (Lepidoptera). Insect Syst Evol. 19(2):117–129.
- Stainton HT. 1856. Descriptions of three species of Indian micro-Lepidoptera. Trans Entomol Soc Lond NS. 3:301–304.