Abstract
Liriope platyphylla is used as an important medicinal plant for fatigue, cough, and inflammation in South Korea. Here, we report the complete chloroplast genome of L. platyphylla. The total genome size of the chloroplast is 157,076 bp with a large single-copy region (LSC: 85,374 bp), a small single-copy region (SSC: 18,748 bp), and inverted repeat regions (IRa and IRb: 26,477 bp). The GC content of the L. platyphylla chloroplast was 37.6%. The cp genome encoded a set of 129 genes, including 83 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The phylogenetic tree analysis indicated that L. platyphylla is closely related to L. spicata.
Liriope platyphylla is an herbaceous plant belonging to the Asparagaceae family and is widely distributed in East Asia (Korea, China, Japan, and Taiwan) (Shin Citation2002). Liriope platyphylla is common in the southern region of South Korea and has been used in traditional medicine for nutritional tonics, expectorants, and diuresis (Han Citation1993). Recent studies of L. platyphylla have reported anti-inflammatory action (Kim et al. Citation2016), immunomodulatory effects (Kim Citation2003), protective effects of the liver (Park et al. Citation2019) and lungs (Lee et al. Citation2011), anti-cancer effects (Wang et al. Citation2013), and memory enhancement effects (Kang and Lee Citation2006).
In this study, we revealed the complete chloroplast sequence of L. platyphylla and phylogenetic information of the species. Fresh leaves (L. platyphylla) were collected from Anyang-myeon, Jangheung-gun, Korea (34°40′1″N, 126°56′42″E). A voucher specimen (TKMII-32) was deposited at the Medicinal Crops Seed Supply Center of the National Institute for Korean Medicine Development (NIKOM). The whole chloroplast DNA was isolated using the DNeasy Plant mini kit (QIAGEN, Hilden, Germany), and the raw read sequence (10,629,338 bp) was achieved by Illumina platform (HiSeq 2500 & NovaSeq) at Genotech Inc. (Yuseong-gu, Daejeon, Korea). Raw reads were assembled via NOVOPlasty v2.6.7 (Dierckxsens et al. Citation2017). The assembled genome was annotated using the Dual Organellar Genome Annotator (Dogma; Wyman et al. Citation2004) and the Chloroplast Genome Annotation, Visualization, Analysis, and GenBank Submission Tool (CPGAVAS2; Shi et al. Citation2019).
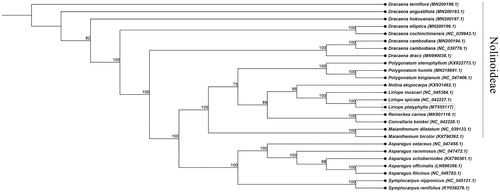
The complete chloroplast genome of L. platyphylla was 157,076 bp in length and consisted of two single-copy regions (large single-copy region (LSC) with 85,374 bp, and small single-copy region (SSC) with 18,748 bp) and a pair of inverted repeat (IR) regions (IRa and IRb with 26,447 bp). The cp genome encoded a total of 129 genes, including 83 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The total GC content of the chloroplast genome was 37.6%, with corresponding contents in the LSC, SSC, and IR regions of 35.6, 31.2, and 43.0%, respectively. The alignment was conducted using MAFFT (Katoh and Standley Citation2013). The phylogenetic tree (neighbor joining tree) was constructed using CLC Main Workbench 7 with a bootstrap set to 10,000 (). The phylogenetic tree indicated that L. platyphylla was most similar to L. spicata.
Acknowledgments
We thank Dr. S. H. Cha and S.W. Lee for assisting with the nucleotide data analysis of plant materials.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data of this study are publicly published in NCBI's GenBank (https://www.ncbi.nlm.nih.gov/genbank/), reference number MT555117.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Han DS. 1993. Pharmacognosy. 5th ed. Seoul, Korea. Dongmyungsa; p. 148.
- Kang YG, Lee TH. 2006. Liriopis Tuber improves stress-induced memorial impairments in rats. Korea J Herbol. 21:63–75.
- Kim H. 2003. Effect of Maekmoondong-Tang on the immunomodulatory action. J Physiol Pathol Korean Med. 17:946–951.
- Kim MJ, Yoo YC, Sung NY, Lee J, Park SR, Shon EJ, Lee BD, Kim MR. 2016. Anti-inflammatory effects of Liriope platyphylla in LPS-stimulated macrophages and endotoxemic mice. Am J Chin Med. 44(6):1127–1143.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lee ES, Yang SY, Kim MH, Namgung U, Park YC. 2011. Effects of root of Liriope Spicata on LPS-induced lung injury. J Physiol Pathol Korean Med. 25:641–649.
- Park G, Parveen A, Kim JE, Cho KH, Kim SY, Park BJ, Song YJ. 2019. Spicatoside A derived from Liriope platyphylla root ethanol extract inhibits hepatitis E virus genotype 3 replication in vitro. Sci Rep. 9(1):1–11.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Shin JS. 2002. Saponin composition of Liriope platyphylla and Ophiopogon japonicas. Korean J Crop Sci. 47(3):236–239.
- Wang HC, Wu CC, Cheng TS, Kuo CY, Tsai YC, Chiang SY, Wong TS, Wu YC, Chang FR. 2013. Active constituents from Liriope platyphylla root against cancer growth in vitro. Evidence-Based Complement Altern Med. 2013:1–10.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.