Abstract
The complete chloroplast genome of an endangered endemic species in China Tilia taishanensis was sequenced with Illumina HiSeq 2000 platform. It was a typical quadruple structure as other plants of Tilia with 162,803 bp in length, including a large single copy (LSC: 91,114 bp) region and a small single copy (SSC: 20,379 bp) which were separated by a pair of inverted repeats (IRa, b: 25,655 bp) region. The overall GC content is 36.5%. A total of 129 genes was annotated which contained 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. ML Phylogenetic analysis compared with 33 expressed chloroplast genomes revealed that T. taishanensis was a sister to other Tilia species.
Tilia taishanensis S. B. Liang is an endangered endemic ornamental, timber and nectar plant in East China which only distributed in Shandong Province with about 500 individuals in total (Shubin Citation1985; Hanbin Citation1990; Wenqing et al. Citation2016; Fazeng et al. Citation2016). Its distribution areas are easily affected by tourism development, grazing and forest tending (Wenqing et al. Citation2016). And which was classified as endangered species according to the IUCN standard (Boqiang et al. Citation2019). The embryology (Hong Citation2014), cuttage breeding of T. taishanensis (Xiaolong et al. Citation2015) had been carried out. However, the understanding of its resource information was still limited. Genome mining would be helpful to the study of endangered mechanism of T. taishanensis and promote its conservation and utilization.
The fresh leaves of T. taishanensis were collected from the living individual permanently conserved in the Tilia gene bank (36.628°N, 117.1636°E) which was grafted from the wild tree. The specimens were preserved in National Plant Specimen Resource Center (http://www.cvh.ac.cn/, barcodes SDF1009895 and SDF1009900). Total genomic DNA was extracted by the Plant DNA extraction Kit (TIANGEN, Beijing, China) according to the requirements of the reagent company. Paired-end reads were constructed according to the Illumina library preparation protocol and sequenced on an Illumina HiSeq 2000 platform. The whole chloroplast genome of T. taishanensis was assembled by MITObim 1.8 (Hahn et al. Citation2013) and wad annotated in DOGMA (http://dogma.ccbb.utexas.edu/). Maximum-likelihood (ML) phylogenetic tree with 100 bootstrap replicates was inferred using TreeBeST 1.9.2 (Vilella et al. Citation2008).
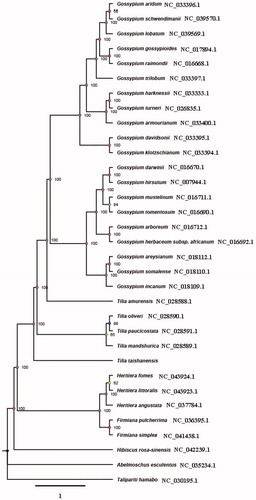
The chloroplast genome of T. taishanensis (GenBank accession number MT591529) was also a typical quadruple structure with 162,803 bp in length that contains a large single copy (LSC: 91,114 bp) region and a small single copy (SSC: 20,379 bp), which were separated by a pair of inverted repeats (IRa, b: 25,655 bp) region. The overall GC content was 36.5%. A total of 129 genes were annotated, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Among these, 7 genes had a single intron, ycf3 and clpP had two introns. The rps12 had four copies, and rrn4.5, rrn5, rrn16, rrn23 had two copies respectively. The chloroplast genome of T. Taishanensis was similar to that of other four species of Tilia, but there were obvious differences. ML Phylogenetic analysis compared with 33 expressed chloroplast genomes revealed that T. taishanensis was a sister to other Tilia species ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/genbank/, GenBank accession number MT591529.
Additional information
Funding
References
- Boqiang T, Lei W, Dekui Z, Ping D, Dan L, Fusheng W, Aizong S,Xiaoman X. 2019. Investigation of endemic tree species resources in Shandong Province. Anhui Agri Sci. 47(16):130–132.
- Fazeng L, Wenqing L, Shoujin F. 2016. Woody flora of Shandong (Volume I and II). Beijing: Science Press.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Hanbin C. 1990. Flora of Shandong (Volume I and II). Qingdao: Qingdao press.
- Hong X. 2014. Embryology of Tilia taishanensis [master’s thesis]. Shandong Agricultural University.
- Shubin L. 1985. Two new species of Tilia from Shandong Province. Journal of Shandong Forestry Science and Technology. 4(2):70.
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. 2008. Ensembl Compara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19 (2):327–335.
- Wenqing L, Dekui Z, Xiaoman X. 2016. Rare and endangered protected tree species in Shandong. Beijing: Science Press.
- Xiaolong W, Wanyuan Y, et al. 2015. Study on cutting propagation technology of Tilia taishanensis. China Agronom Bull. 31(34):16–19.