Abstract
Michelia compressa is an evergreen ornamental tree species. The high-throughput sequencing technology was used to sequence and assemble the chloroplast genome of Michelia compressa. Results showed that the chloroplast genome is 160,061 bp in length, of which the inverted repeats sequence (IRs) is 26,581 bp, the large single-copy region (LSC) and the small single copy region (SSC) are 88,097 bp and 18,802 bp, respectively. The GC content of the plastome was 39.2%, with 43.2%, 37.9% and 34.2% in IRs, LSC and SSC, respectively. A total of 132 genes are annotated, 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. This study enriched the Michelia compressa genomic information which provides the basis for rational exploitation and utilization of germplasm resources.
Magnoliaceae is one of the primitive groups of angiosperms which contains 16 genera that more than 300 plants. Among that Michelia as the second largest genus of the family magnoliaceae, consists of about 50 species (Keng Citation1996), and there are 38 species in China, living mainly from southwest to southeast. The flower buds of this plant are always used as traditional Chinese medicine, which have been used for treating laryngitis, rhinitis, inflammation. But, at present, there are still great differences in the division of the position of the magnoliaceae, especially the subspecies of the family. Chloroplast has been a valuable tool to be used for phylogenetic studies due to its gene conservation and the lack of recombination (Ravi et al. Citation2008; Lin et al. Citation2012). Here, we reported the complete Chloroplast Genome of Michelia compressa, which provide valuable information for molecular breeding, genetic modification, and phylogenetic study in Magnoliaceae.
The sample of M. compressa was collected from Nanjing Botanical Garden MEM. Sun Yat-Sen, Jiangsu, China (32°02′N, 118°28′E). The voucher specimens were stored in the Institute of Botany, Chinese Academy of Sciences, Jiangsu Province (voucher No.320102190105002LY). Total genomic DNA was extracted from the fresh mature leaves of M. compressa and sequenced on an Illumina Hiseq Xten platform (San Diego, CA). Genome sequences were screened out and assembled with Novoplasty v 2.7.2 (Dierckxsens et al. Citation2017).
The sequence of M. compressa genome has been deposited in Genbank (accession number: MN604380). The total length of M. compressa chloroplast genome was determined to be 160,061 bp in length, of which the inverted repeats sequence (IRs) is 26,581 bp, the large single-copy region(LSC) and the small single copy region (SSC) are 88,097 bp and 18,802 bp, respectively. The GC content of the plastome was 39.2%, with 43.2%, 37.9% and 34.2% in IRs, LSC and SSC, respectively. The genome is structured with 132 genes, including 86 coding protein genes, 37 tRNA genes and 8 rRNA genes. Among these genes, 18 genes were repeated IR regions, including 7 protein-coding genes, 7 tRNA genes and 4 rRNA genes.
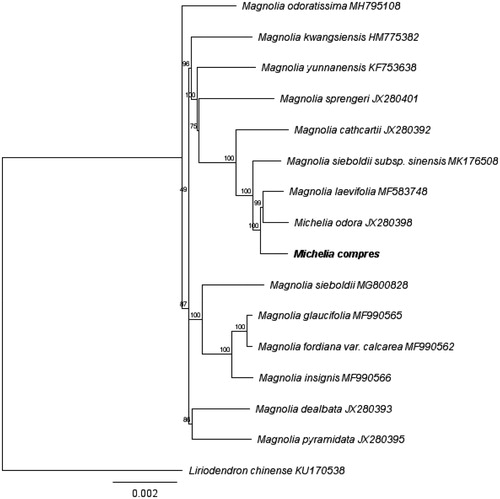
A molecular phylogenetic tree was constructed with MEGA 6.0 (Tamura et al. Citation2013) based on the maximum likelihood method using a dataset of the complete genome sequence of M. compressa with those of 14 individuals of Magnoliaceae obtained from Genbank. The result revealed that Michelia was a polyphyletic genus. M. compressa was closely related with Mangolia laevifolia and Michelia odora, and nested within the other Magnolia species ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank (https://www.ncbi.nlm.nih.gov) with the accession number is MN604380.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Keng H. 1996. Magnoliaceae in flora of Taiwan. 2nd ed. Taipei, Taiwan: Editorial Committee of the Flora of Taiwan.
- Lin CP, Wu CS, Huang YY, Chaw SM. 2012. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol Evol. 4(3):374–381.
- Ravi V, Khurana J, Tyagi A, Khurana P. 2008. An update on chloroplast genomes. Plant Syst Evol. 271(1–2):101–122.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.