Abstract
The species Metidiocerus sp. belonging to the subfamily Idiocerinae (Hemiptera, Cicadellidae). Here, we sequenced and annotated the mitochondrial genome (mitogenome) of Metidiocerus sp. This mitogenome was 15,079 bp long and encoded 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and 2 ribosomal RNA unit genes (rRNAs), and one non-coding region. The nucleotide composition biases toward A and T, which together made up 77.4% of the entirety. All 13 PCGs were initiated by the ATN (ATG, ATT, ATA, and ATC) codon. All PCGs terminate with the stop codons TAA except for COX2, ND4, and ND1 ended with single T. A phylogenetic tree generated by the Bayesian method showed that Metidiocerus sp. is closely related to Idiocerus salicis and Idiocerus herrichii which enriched the mitochondrial genome data of Idiocerinae.
Idiocerinae is one of the largest groups of arboreal leafhoppers, comprising 105 genera and approximately 800 described species (Zhang and Webb Citation2019). The genus Metidiocerus was originally described by Ossiannilsson (Citation1981) containing six species and distributed only in the Palearctic region. Kwon (Citation1985) considered Metidiocerus as a subgenus of Idiocerus and described one new species, Idiocerus (Metidiocerus) nigrolineatus. Metidiocerus was elevated to generic status (Isaev Citation1988). This genus is characterized morphologically by having pair of dark brown longitudinal marks on face, lateral margins of genae slightly concave or less straight at apex, forewing with three subapical cells, hind femur with 2 + 0 apical setal formula, apophysis of style bearing 1–2 setae distally, male dorsal basal abdominal apodemes shorter than ventral ones (Ossiannilsson Citation1981; Kwon Citation1985).
In this study, we sequenced and annotated the complete mitochondrial DNA of the Metidiocerus sp. (MT554451) for the first time. The specimen of Metidiocerus sp. was collected in Shawan County, Xinjiang Uygur Autonomous Region, China in July 2019. The samples and voucher specimens (No. IMNU2019070603) were stored in 100% ethanol in the field and then stored at −30 °C in Inner Mongolia Normal University, China. The entire body without abdomen was shipped to Tsingke (Beijing, China) for genomic extraction. The mitogenome sequence of Metidiocerus sp. was generated using Illumina HiSeq 2000 Sequencing System. De novo assembly of clean reads was performed using SPAdes version 3.11.0 (Bankevich et al. Citation2012). The base composition was analyzed by MEGA version 7 (Kumar et al. Citation2016). Genes were annotated with the MITOS (Bernt et al. Citation2013) web server.
The mitogenome of Metidiocerus sp. consists of a 15,079 bp circular DNA molecule, with 42.1% A, 35.3% T, 12.3% C, and 10.3% G, which has an A/T bias (77.4%, A + T content). It contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and 2 ribosomal RNA unit genes (rRNAs), and one non-coding region. Most genes are encoded on heavy strand except for four PCGs (ND4, ND4L, ND5, and ND1) and eight tRNA genes (tRNA-Gln, Cys, Tyr, Phe, His, Pro, Leu, and Val). All 13 PCGs were initiated by the ATN (ATG, ATT, ATA, and ATC) codon. All PCGs terminate with the stop codons TAA and TAG except for COX2, ND4, and ND1 ended with single T. The 12S rRNA and 16S rRNA genes in the Metidiocerus sp. mitogenome were 751 and 640 bp in size. The sequenced partial non-coding region was 739 bp in length, located after the 12S rRNA.
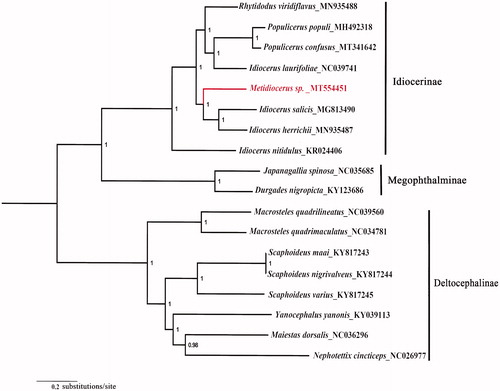
All 13 PCGs sequences were extracted from the mitochondrial DNA sequences of 18 related taxa of Cicadellidae. The concatenated PCGs using Bayesian inference (BI) method in MrBayes version 3.2.1 (Ronquist and Huelsenbeck Citation2003; Ronquist et al. Citation2012) under the GTR + G model. The phylogenetic tree () showed that Metidiocerus sp. is closely related to Idiocerus salicis and Idiocerus herrichii which enriched the mitochondrial genome data of Idiocerinae. The nearly complete mitogenome of Metidiocerus sp. could provide important information for the further studies of Metidiocerus phylogeny.
Figure 1. Phylogenetic tree (midpoint rooted) of the relationships among 18 species of Cicadellidae based on the nucleotide dataset of 13 PCGs. Numbers above the nodes indicate the posterior probabilities of Bayesian inference using MrBayes version 3.2.1 under the GTR + G model. Branch lengths represent means of the posterior distribution. The GenBank numbers of all species are shown in the figure.
Acknowledgments
We thank Dr. Xianguang Guo (Chengdu Institute of Biology, Chinese Academy of Sciences) for his assistance in analyzing the data and revising the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s). The authors alone are responsible for the content and writing of this article.
Data availability statement
The authors confirm that the data supporting the finding of this study are available within its supplementary material. https://www.ncbi.nlm.nih.gov/nuccore/MT554451.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov AS, Lesin V, Nikolenko S, Pham S, Prjibelski A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Isaev VV. 1988. Megipocerini, new tribe of leafhoppers (Homoptera, Cicadellidae, Idiocerinae). Entomotaxonomia. 10(1–2):65–69.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Kwon YJ. 1985. Classification of the leaghopper-pests of the subfamily idiocerinae from Korea. The Korean. J Entomol. 15(1):61–73.
- Ossiannilsson F. 1981. The Auchenorrhyncha (Homoptera) of Fennoscandia and Denmark. Fauna. Ent Scand. 7:223–593.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Zhang B, Webb MD. 2019. A checklist and key to the idiocerine leafhoppers (Hemiptera: Cicadellidae) of Hainan Island, with description of a new genus and new species. Zootaxa. 4576(3):581.
