Abstract
In this study, we firstly reported the complete mitochondrial genome of Populicerus confuses. The complete mitochondrial genome was 16,395 bp in length which overall base composition was 41.43% A, 36.30% T, 11.54% C, and 10.73% G. It consisted of 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes (12S and 16S rRNA), and a control region (D-loop region). The complete mitochondrial genomes of P. confuses and other 9 species were used for phylogenetic analysis using the Bayesian method. The resulting phylogenetic tree confirms that the Populicerus populi is most closely related to P. confuses. The mitogenome provided the valuable evidence on phylogenetic relationship of the Idiocerinae at the molecular level.
Cicadellidae is a common component of all zoogeographic regions, with comprises 22,000 species in the world (Camisão et al. Citation2014). Populicerus confuses is a member of Idiocerinae subfamily. Idiocerinae is a moderately large, diverse group of arboreal leafhoppers, for which about 107 genera and 800 species are recorded worldwide (Zhang and Webb Citation2019). Currently, elucidating the structure of P. confusus mitogenome is important for understanding its diversity and evolution. The results will contribute for the identification and further phylogenetic analyses of the members of Idiocerinae.
In this study, we sequenced and analyzed the mitogenome of P. confuses, with the number of GenBank MT341642. The specimen was collected from Norjinkhairkhan, Khovd Province, Mongolia (N47.99, E91.62) on August 2019, and was deposited in the insect specimen room of Research Institute of Inner Mongolia Normal University (number IMNU2019081208). Genomic DNA isolated was sequenced using Illumina NovaSeq. The entire body without abdomen was shipped to Genepioneer (Nanjing, China) for genomic extraction. Sequencing was performed on an Illumina HiSeq 2000 instrument. The resultant reads were assembled using the SPAdes v3.10.1 (Bankevich et al. Citation2012). The complete mitochondrial genome was annotated with MITOS (Bernt et al. Citation2013). All tRNA genes were identified by tRNAscan-SE Search Server (Lowe and Chan Citation2016).
The complete mitochondrial genome of P. confuses was 16,395 bp in length with the A + T content of 77.73% (T 36.30%, C 11.54%, A 41.43%, and G 10.73%), which is within the range reported for hemipteran mitogenomes (68.86–86.33%; Zhang et al. Citation2014). It consisted of 13 PCGs (COX1-3, ND1-6, ND4L, ATP6, ATP8, and Cytb), 22 tRNA genes, 2 rRNA genes, and 1 D-loop region. Among the 13 PCGs, all genes took the start codon of ATN, while ATP8 and ND5 got TTG. The termination codon of these PCGs had two types (10 genes were TAA and 3 genes were single T––). In 13 PCGs, most genes are encoded on the H-strand, except for ND4, ND4L, ND5, and ND1. Eight tRNA genes (tRNA-Gln, Cys, Tyr, Phe, His, Pro, Leu1 and Val) are encoded on the L-strand. The 16S rRNA, with a length of 635 bp, is located between tRNA-Val and tRNA-Leu, and the 12S rRNA, with a length of 738 bp, is located between tRNA-Val and D-loop. The control region, with a length of 2107 bp, is located after 12S rRNA.
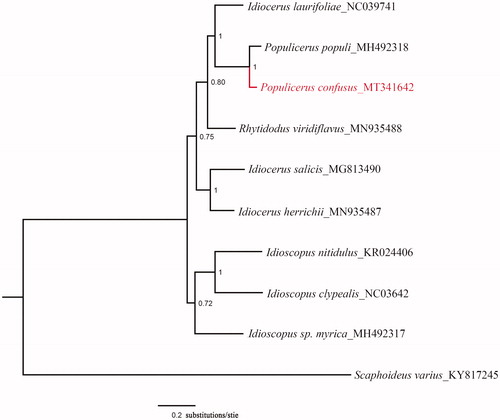
Phylogenetic analysis was carried out on the basis of 10 available mitogenomes of Cicadellidae insects in GenBank. It inferred from Bayesian inference using MrBayes v.3.2.1 (Ronquist and Huelsenbeck Citation2003; Ronquist et al. Citation2012) based on the PCGs of nine species of Idiocerinae and one outgroup. Sequences were aligned with MEGA6 software (Tamura et al. Citation2013). The phylogenetic tree showed that Populicerus populi is most closely related to P. confuses (). The mitogenome provided the valuable evidence on phylogenetic relationship of the Idiocerinae at the molecular level.
Acknowledgements
We would like to express our sincere gratitude to Dr. Xianguang Guo (Chengdu Institute of Biology, Chinese Academy of Sciences) for his helpful suggestions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors confirm that the data supporting the finding of this study are available within its supplementary material. https://www.ncbi.nlm.nih.gov/nuccore/MT341642
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov AS, Lesin V, Nikolenko S, Pham S, Prjibelski A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Camisão BM, Cavichioli RR, Takiya DM. 2014. Eight new species of Oragua Melichar, 1926 (Insecta: Hemiptera: Cicadellidae) from Amazonas State, Brazil, with description of the female terminalia of Oragua jurua Young, 1977, and new records for the genus. Zootaxa. 3841(4):501–527.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Zhang B, Webb MD. 2019. A checklist and key to the idiocerine leafhoppers (Hemiptera: Cicadellidae) of Hainan Island, with description of a new genus and new species. Zootaxa. 4576(3):587.
- Zhang KJ, Zhu WC, Rong X, Liu J, Ding XL, Hong XY. 2014. The complete mitochondrial genome sequence of Sogatella furcifera (Horváth) and a comparative mitogenomic analysis of three predominant rice planthoppers. Gene. 533(1):100–109.