Abstract
The chloroplast genome sequence of Michelia martini was sequenced using high-throughput sequencing technology and analyzed phylogenetically in the present study. The complete chloroplast genome was 159,819 bp in length, including a large single copy region (LSC) of 88,078 bp and small single copy region (SSC) of 18,801 bp, and a pair of inverted repeat regions (IR) of 26,470 bp. The contents of CG in the chloroplast genome were 39.29%. The sequence contained 128 unique genes, including 81 protein-coding genes, 38 tRNA genes and 8 rRNA genes. The phylogenetic analysis revealed that M. martini is closely related to Magnolia maudiae.
Michelia martini Finet & Gagnepain ex H. Léveillé is an excellent broad-leaved tree species for landscaping and wood production belonging to the Magnoliaceae family. The Magnoliaceae is considered as one of the most primitive groups of angiosperms (Li and Guo Citation2014). Magnolioideae is thought to include one genus Magnolia (Nooteboom Citation2000; Figlar and Nooteboom Citation2004) or divided into several smaller genera (Law Citation1984; Liu Citation2004; Xia et al. Citation2009). The flowering period is from February to march, and the fruit period is from September to October, it also had certain medicinal value of the essential oil from fresh leaves which had significant effects on lowering blood pressure, cough suppression, expectorant and antifungal effects (Lei et al. Citation2016). However, there has been no genomic studies on M. martini. Chloroplast has been a valuable tool to be used for phylogenetic studies due to its gene conservation and the lack of recombination (Ravi et al. Citation2008; Lin et al. Citation2012). Here, the complete cp genome sequence of M. martini was assembled and analyzed using high-throughput sequencing technology, the complete cp genome (GenBank accession number MT44441) to retrieve valuable resources for the conservation and phylogeny of M. martini.
The fresh leaves were collected from three adult M. martini plants from that were growing in Longzhong Botanical Garden (32°10′N, 112°10′E), Hubei, China. The specimen of M. martini was stored in the herbarium from the Three Gorges University, the accession number was 00126151. The leaves were stored at −80 °C until used. Total DNA was extracted to construct a library for sequencing with Illumina Hiseq 2500 platform (Illumina, San Diego, CA, USA). Additionally, MITObim v 1.8 (https://github.com/chrishah/MITObim) was used to assemble the complete circular cp genome sequence (Hahn et al. Citation2013). The cp genome was annotated and manually adjusted with CpGAVAS (Liu et al. Citation2012). the circular plastid genome map was completed with the help of the online program OrganellarGenome DRAW (OGDRAW) (Lohse et al. Citation2013) and the annotated sequence was submitted to NCBI.
The total length of the chloroplast genome was 159,819 bp presenting a typical quadripartite structure, of which the length of a large single copy region (LSC) was 88,078 bp and the length of a small single copy (SSC) region was 18,801 bp, which were separated by the IRA and IRB of 26,470 bp. The contents of CG in the chloroplast genome was 39.29%. A total of 128 chloroplast genes were annotated, containing 81 protein-coding genes (63.28%), 38 transporter RNA genes (29.69%), eight ribosomal RNA genes (6.55%) , one pseudogene (0.78%) was inferred to be pseudogenes. Ten protein-coding genes (rps12, rpl2, trnV-UAC, trnL-UAA, atpF, ycf68, trnI-GAU, ndhA, trnA-UGC and ndhB) contained one intron, while clpP and ycf3 each contained two introns.
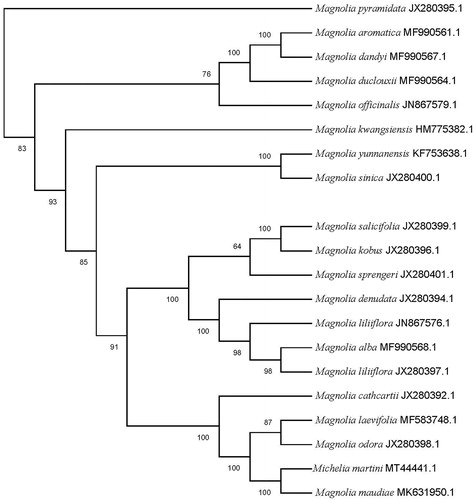
A characterization of the genetic relationship between M. martini and related taxa may provide valuable information for broadening the genetic basis of rootstock breeding programs. Thus, we further investigated the phylogenetic relationships between the chloroplast genome sequences of M. martini and 19 other species of Magnoliaceae reported in Genbank of NCBI database using MEGA 7.0 (Kumar et al. Citation2016) (https://www.megasoftware.net). The phylogenetic analysis showed that M. martini was closely related with Magnolia maudiae, forming a clade included in Magnolia (). The cp genome of M. martini will provide useful genomic resources for further study on genetic diversity and conservation of this species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov/, reference number MT44441.
Additional information
Funding
References
- Figlar RB, Nooteboom HP. 2004. Notes on Magnoliaceae IV. Blum. J Plant Tax Plant Geog. 49(1):87–100.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-abaiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Law YW. 1984. A preliminary study on the taxonomy of the family Magnoliaceae. Acta Phytotaxon. Sin. 22(2):89–109.
- Lei LH, Yu XY, Li YL. 2016. Analysis of volatile oil components from fresh twigs of Michelia martini Levl. in four seasons. J Hunan Agri Univ Nat Sci Edn. 42(3):296–300.
- Li ZY, Guo R. 2014. Review on propagation biology and analysis on endangered factors of endangered species of Manglietia. Life Sci Res. 18:90–94.
- Lin CP, Wu CS, Huang YY, Chaw SM. 2012. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol Evol. 4(3):374–381.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13(1):715.
- Liu YH. 2004. Woonyoungia Law. In Liu Yuhu (ed.) Magnolias of China. Beijing: Beijing Science and Technology Press. p. 381–383.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–58.
- Nooteboom HP. 2000. Different looks at the classification of the Magnoliaceae. Proceedings of the International Symposium on the Family Magnoliaceae. Beijing: Science Press. p. 26–37.
- Ravi V, Khurana J, Tyagi A, Khurana P. 2008. An update on chloroplast genomes. Plant Syst Evol. 271(1-2):101–122.
- Xia NH, Liu YH, Nooteboom HP. 2009. Magnoliaceae. In: Xia NH, Liu YH, Nooteboom HP (Eds.) Flora of China. Vol. 7. Beijing: Missouri Botanical Garden Press and Science Press. p. 48.