Abstract
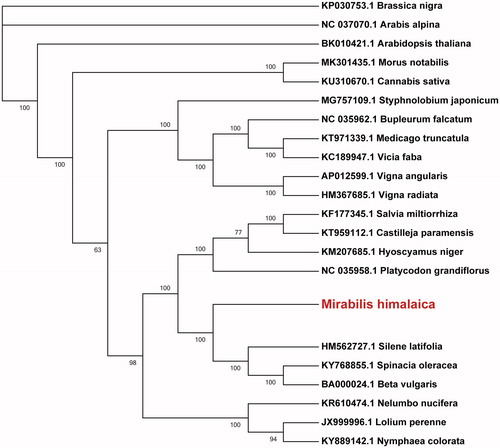
The complete mitochondrial genome of the important medicinal plant Mirabilis himalaica has been reported in this study. It was 346,363 bp in size with a GC content of 44.6% and contained 42 protein-coding genes (30 known functional genes, 11 ORF genes and one ycf2 gene of unknown function), 27 tRNA genes, and four rRNA genes. Most of the genes were single-copy genes, only three were duplicated and two were multi-copy. Phylogenetic analysis showed M. himalaica to be closely clustered with Silene latifolia, Spinacia oleracea and Beta vulgaris, all species belong to the order Caryophyllales.
Mitochondrial genome is a single circular molecule containing genes that are inherited together (Berlin and Ellegren Citation2001), therefore carries consistent phylogenetic information and has been exploited for the reconstruction of robust phylogenies in different taxa (Yu et al. Citation2018; Zhang et al. Citation2020). Mirabilis himalaica (Edgew.) Heimerl is an important medicinal plant species of the family Nyctaginaceae, but its exact taxonomic position is controversial (Wang et al. 2018). Unfortunately, there is no report of mitochondrial genome sequence of species in Nyctaginaceae and only single-gene molecular markers were used to study the taxonomic status of M. himalaica (Wang et al. 2018). To circumvent these limitations, we report here the complete mitochondrial genome of M. himalaica (Genbank accession number: MT535663), the first of the family Nyctaginaceae.
Genomic DNA was obtained from fresh leaves of M. himalaica collected from Nyingchi City (N: 29°40′23.40″, E: 94°20′28.28″), Tibet Autonomous Region, China, in August 2019 using the CTAB method (Doyle and Doyle Citation1987). The voucher specimen was deposited at Tibet Agriculture and Animal Husbandry University (Accession number: ZY19082301). The mitochondrial genome sequencing was performed at Wuhan Bio-mall Biotechnology Co., Ltd, Wuhan, China, using the Illumina Hiseq sequencing technology (Illumina, San Diego, CA).
The generated 150 bp paired-end reads were quality-checked using the Fastqc tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and low-quality sequences were filtered out. SPAdes v3.9.0 (Nurk et al. Citation2013) and MITOS WebServer (http://mitos.bioinf.uni-leipzig.de) were used to assemble and annotate the mitochondrial DNA sequences. tRNA sequences were submitted to the tRNAscan-SE v1.21 online website (http://lowelab.ucsc.edu/tRNA Scan-SE/) and rRNA sequences were submitted to RNAmmer 1.2 Server (http://www.cbs.dtu.dk/services/RNAmmer/). Phylogenetic tree that shows the comprehensive evolutionary relationship with 21 other species was built in MEGA 7 based on maximum likelihood (ML) (Kumar et al. Citation2016).
The assembled mitochondrial genome was a closed circular molecule with 346,363 bp in length, representing an average sequencing depth of 171X. It encoded 42 protein-coding genes, including nine NADH dehydrogenase (nad1, 2, 3, 4, 4L, 5, 6, 7, 9), three cytochrome c oxidase (cox1, 2, 3), five cytochrome c biogenesis (ccmB, C, C, FN, FC), one succinate dehydrogenase (sdh3), one apocytochrome b (cob), five ATP synthase (atp1, 4, 6, 8, 9), four ribosomal proteins (rps3, 4, 12, 13), one maturase (matR), one membrane transporter (tatC), 11 ORF (ORF223, 223, 243, 277, 279, 284, 287, 288, 312, 341, 399) and one ycf2 gene of unknown function. In addition, the genome contained four rRNA genes (rrn5, 18, 26, 26) and 27 tRNA genes (trnC-GCA, trnC-GCA, trnC-GCA, trnD-GTC, trnE-TTC, trnF-GAA, trnG-GCC, trnH-GTG, trnK-TTT, trnM-CAT, trnM-CAT, trnM-CAT, trnM-CAT, trnM-CAT, trnM-CAT, trnM-CAT, trnM-CAT, trnN-GTT, trnN-GTT, trnP-TGG, trnQ-TTG, trnQ-TTG, trnS-GCT, trnS-GGA, trnS-TGA, trnW-CCA, trnY-GTA). The genes nad1, 2, 5 were trans-spliced. Most of the genes were single-copy genes, only three were duplicated and two were multi-copy. The overall base composition of the M. himalaica mitochondrial genome was A: 27.8%, G: 22.4%, C: 22.2%, T: 27.6%, and the GC content is 44.6%.
To confirm the phylogenetic position of M. himalaica, we constructed ML tree with 21 other species (). Mirabilis himalaica was found closely clustered with Silene latifolia (Caryophyllaceae), Spinacia oleracea (Chenopodiaceae) and Beta vulgaris (Amaranthaceae), all belonging to the order Caryophyllales (Gruzdev et al. Citation2018). This confirms the phylogenetic placement of M. himalaica in the order Caryophyllales. In future, we will report mitochondrial genome sequences of related species in the family Nyctaginaceae to clarify the taxonomic status of M. himalaica.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore, reference number MT535663.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Berlin S, Ellegren H. 2001. Evolutionary genetics. Clonal inheritance of avian mitochondrial DNA. Nature. 413(6851):37–38.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gruzdev EV, Mardanov AV, Beletsky AV, Ravin NV, Skryabin KG. 2018. The complete mitochondrial genome of the carnivorous flowering plant Nepenthes X Ventrata. Mitochondrial DNA Part B. 3(2):1259–1260.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, Prjibelsky A, Pyshkin A, Sirotkin A, Sirotkin Y, et al. 2013. Assembling Genomes and Mini-metagenomes from Highly Chimeric Reads. In: Deng M, Jiang R, Sun F, Zhang X, editors. Research in computational molecular biology. RECOMB 2013. Lecture Notes in Computer Science, vol 7821: 158–170. Berlin, Heidelberg: Springer.
- Yu T, Sun L, Cui H, Liu S, Men J, Chen S, Chen Y, Lu C. 2018. The complete mitochondrial genome of a tertiary relict evergreen woody plant Ammopiptanthus mongolicus. Mitochondrial DNA Part B. 3(1):9–11.
- Zhang J, Zhang S, Zhang Y. 2020. The complete mitochondrial genome of a mangrove plant: Aegiceras corniculatum and its phylogenetic implications. Mitochondrial DNA Part B. 5(2):1502–1503.