Abstract
Endiandra hainanensis Merr. et Metc. ex Allen is a woody plant belonging to the genus Endiandra R. Brown Prodr, in the family Lauraceae. To better determine its phylogenetic location with respect to the other Endiandra species, the complete plastid genome of E. hainanensis was sequenced. The whole plastome size is 158,558 bp that includes a pair of inverted repeat (IR) regions of 25,513 bp, one large single-copy (LSC) region of 89,285 bp, and one small single-copy (SSC) region of 18,247 bp. The overall GC content of the whole plastome is 39.04%. Maximum likelihood phylogenetic analysis with TVM + F + R2 model was conducted using 20 complete plastomes of the Lauraceae, which supports the close relationship between E. hainanensis and E. dolichocarpa.
Endiandra hainanensis Merr. et Metc. ex Allen, a tree species distributed in Asian (Rohwer Citation1993), was assigned to the genus Endiandra in the family Lauraceae (Editorial Committee of Flora of China, Citation1989–2013). There are about 30 species in the genus Endiandra, three of which, namely E. hainanensis, E. dolichocarpa, and E. coriacea, are distributed in China (Editorial Committee of Flora of China, Citation1989–2013). Several species of Endiandra play a role in the commercial timber industry and crucially constitutes tree species of evergreen broad-leaved forest in the tropical and subtropical areas as well (Göltenboth et al. Citation2006). For a better understanding of the relationships between E. hainanensis and other Endiandra species, it is necessary to reconstruct a phylogenetic tree based on high-throughput sequencing approach.
Fresh leaves of E. hainanensis in Mengla County (Xishuangbanna, Yunnan, China, Long. 101.3428 E, Lat. 21.3645 N, 775 m) were collected for DNA extraction (Doyle and Dickson Citation1987). The voucher was deposited at the Biodiversity Research Group of Xishuangbanna Tropical Botanical Garden (Accession Number: XTBG-BRG-SY36739). The whole plastid genome was sequenced following Zhang et al. (Citation2016), and 15 universal primer pairs were used to perform long-range PCR for next-generation sequencing. The contigs were aligned using the publicly available plastid genome of E. hainanensis (Accession number LAU00154) (Song et al. Citation2019) and annotated in Geneious 11.1.4.
The plastome of E. hainanensis (LAU00154), with a length of 158,558 bp, was 52, 13 bp smaller than that of E. dolichocarpa (158,610 bp, LAU00053) and E. muelleri (158,571 bp, LAU00054). The lengths of the inverted repeats (IRs), large single-copy (LSC), and small single-copy (SSC) regions of E. hainanensis were 25,513, 89,285, and 18,247 bp, respectively. The overall G + C content was 39.04%.
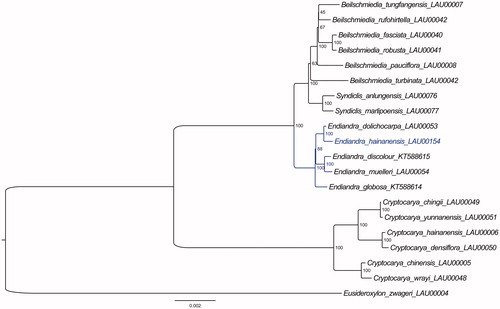
In addition, based on the 20 published plastid genome sequences (Song et al. Citation2017), we reconstructed a phylogenetic tree () to confirm the evolutionary relationship between E. hainanensi and other related species with published plastomes in Beilschmiedia, Cryptocarya, Syndiclis, and Eusideroxylon species as outgroup. Maximum likelihood (ML) phylogenetic analyses were performed based on TVM + F + R2 model in the iqtree version 1.6.12 program with 1000 bootstrap replicates (Nguyen et al. Citation2015). The ML phylogenetic tree with 45–100% bootstrap values at each node supports the fact that Endiandra species can be grouped into three clades, and that sisterhood of E. hainanensis and E. dolichocarpa, with E. discolor and E. muelleri being the next sister group, followed by E. globosa ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The chloroplast genome data of the E. hainanensis has been already submitted to Chloroplast Genome Database (https://lcgdb.wordpress.com). The accession number is LAU00154.
References
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon. 36(4):715–722.
- Editorial Committee of Flora of China. 1989–2013. Flora of China. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press.
- Göltenboth F, Timotius KH, Milan PP, Margraf J. 2006. Ecology of insular Southeast Asia: the Indonesian Archipelago. Amsterdam: Elsevier; p. 320–321.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Rohwer JG. 1993. Lauraceae. Vol. 2. In: Kubitzki K, Rohwer JG, Bittrich V, editors. The families and genera of vascular plants. Berlin: Springer-Verlag; p. 385.
- Song Y, Yu WB, Tan YH, Liu B, Yao X, Jin JJ, Padmanaba M, Yang JB, Corlett RT. 2017. Evolutionary comparisons of the chloroplast genome in Lauraceae and insights into loss events in the Magnoliids. Genome Biol Evol. 9(9):2354–2364.
- Song Y, Yu WB, Tan YH, Jin JJ, Wang B, Yang JB, Liu B, Corlett RT. 2019. Plastid phylogenmics improve phylogenetic resolution in the Lauraceae (Accepted). doi: 10.1111/jse.12536.
- Zhang T, Zeng CX, Yang JB, Li HT, Li DZ. 2016. Fifteen novel universal primer pairs for sequencing whole chloroplast genomes and a primer pair for nuclear ribosomal DNAs. J Syst Evol. 54(3):219–227.