Abstract
The complete mitochondrial genome of Pseudocolochirus violaceus was obtained and described in this study. This complete mitochondrial genome is 15,752 bp in length and consists of 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes. Except ND6 and 5 tRNAs, the others were encoded by the heavy strand. The overall base composition of the heavy-strand was 38.65% A, 12.09% G, 24.31% C, and 24.95% T, with a high A + T content of 63.6%. The phylogenetic analysis suggested that P. violaceus was closest to Cucumaria miniata. The newly described mitochondrial genome may provide valuable data for phylogenetic analysis for Holothuroidea.
Holothuroidea, as known as sea cucumber, is one of five echinoderms. It includes more than 900 described species in China (Liao Citation1997). As seafood, sea cucumbers are usually processed into a dried product known as ‘beche-de-mer’. They are regarded as traditional medicine, delicacies, and aphrodisiacs in Asia (Jaquemet and Conand Citation1999). Pseudocolochirus violaceus, also named sea apple cucumber, is a member of cucumaria. It is naturally distributed in the Indo-Pacific region, especially in Beibu Gulf of China, Sri Lanka, Vietnam, the red sea and Australia, and so on (Liao Citation1997). Pseudocolochirus violaceus was found mostly in sand-mud layer from 18 to 70 m deep in sea (up to 100 m) (Liao Citation1997). Dolmatov et al. (Citation2012) found that P. violaceus was able to regenerate the anterior part of the body. Artificial breeding and larval rearing of P. violaceus was reported (Ajith Kumara et al. Citation2016). Unfortunately, only two DNA sequences (16 and 12 s) of P. violaceus could be found in GenBank. Lack of genetic resources has hindered conservation and utilization of P. violaceus. In this study, the complete mitochondrial genome of P. violaceus and its phylogenetic relationships within other sea cucumbers were investigated.
The specimen was collected from Shenzhen, Guangdong province, China (N22°35′, E114°31′) and kept in the Zoological Herbarium, Lingnan Normal University(Acc. Number SC20200319-3). The dorsal body muscles of P. violaceus were fixed in 100% ethanol and stored at −20 °C. Approximately 30 mg of muscle tissue was used for mitochondrial DNA(mtDNA) extraction with TIANamp Marine Animals DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s specification. MtDNA was sequenced using the Illumina Hiseq Sequencing System (Illumina, San Diego, CA, USA). The clean data were acquired and assembled by the SPAdes and PRICE (Bankevich et al. Citation2012). BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) and ORFs finder (https://www.ncbi.nlm.nih.gov/orffinder/) were used to identify and annotated protein-coding genes (PCGs). tRNAscan-SE 2.0 (Lowe and Chan Citation2016) was used to identified tRNA genes. A phylogenetic tree was constructed using MEGA 6.0 software.
The mitochondrial genome of P. violaceus is 15,752 bp in length (GenBank accession number: MT587564) and containing the typical set of 13 PCGs, 22 tRNA and 2 rRNA genes, and one putative control regions. The heavy strand consists of 38.65% A, 12.09% G, 24.31% C and 24.95% T bases, with a high A + T content of 63.60%. Of the 37 genes, 6 were encoded by the light strand, and 31 were encoded by the heavy strand. Eleven PCGs were initiated by ATG, and ND2 was started with GTG. Nine PCGs were terminate with the TAA as stop codon, while cox1, ND4 and ND6 were terminate with the TAG. Twenty-two tRNA genes, ranged in size from 65 to 73 bp.
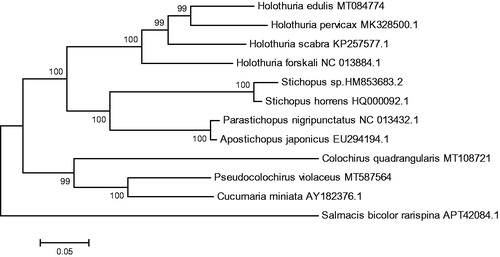
The phylogenetic tree was constructed based on 13 concatenated PCGs from 11 sea cucumbers from GenBank database, by maximum likelihood (ML) method. Salmacis bicolor rarispina was used as an outgroup for tree rooting (). It was demonstrated that P. violaceus was clustered with Cucumaria miniata firstly and both of them were clustered with Colochirus quadrangularis. This result suggested that the P.violaceus is closely related to C. miniata and C. quadrangularis. This newly reported genome of P. violaceus will contribute to future phylogenetic studies and population genetic analyses for P. violaceus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number MT587564.
Additional information
Funding
References
- Ajith Kumara PAD, Jayanatha JS, Pushpakumara J, Bandara W, Dissanayake D. 2016. Artificial breeding and larval rearing of three tropical sea cucumber species – Holothuria scabra, Pseudocolochirus violaceus and Colochirus quadrangularis – in Sri Lanka. SPC Beche-de-Mer Infor Bul. 33:30–37.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Dolmatov IY, Khang NA, Kamenev YO. 2012. Asexual reproduction, evisceration, and regeneration in holothurians (Holothuroidea) from Nha Trang Bay of the South China Sea. Russ J Mar Biol. 38(3):243–252.
- Jaquemet S, Conand C. 1999. The beche-de-mer trade in 1995/1996 and an assessment of exchanges between the main world markets. SPC Beche-de-Mer Info Bull. 12:11–14.
- Liao Y. 1997. Fauna sincia: phylum Echinodermata class Holothuroidea. Beijing: Science Press.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: Search and contextual analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–57.