Abstract
We sequenced and annotated the species of Acroneuria carolinensis which is the second species representative of the genus Acroneuria to provide mitochondrial genome data for order Plecoptera. The complete mitochondrial genome of A. carolinensis was 15,718 bp in length and possessed 22 transfer RNA (tRNA) genes, 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes and one control region. The A + T account of the whole genome, PCGs, tRNAs, rRNAs, and the control region was 64.6%, 62.7%, 67.8%, 68.6%, and 73.9%, respectively. Ten PCGs used the normal start codon except ND2, ND5, and ND1 used the exceptional GTG and TTG as start codon. All PCGs stopped with the terminal codon TAN, but only COII gene used the single T––. The phylogenetic tree was based on the PCG12R matrix (including PCGs first and second codons and two rRNAs) by Bayesian inference (BI) method and we found that the genus Acroneuria and Sinacroneuria were clustered in a clade.
For family Perlidae, there are more than 1100 species over the world at present, and mostly are distributed in the north of the earth while a few extend to southern Africa and South America (DeWalt et al. Citation2020). The genus Acroneuria is belonged to the subfamily Acroneuriinae which currently has 36 valid species, but only seven species are distributed in China (Gutiérrez-Fonseca and Springer Citation2011; Murányi and Li Citation2016; Chen and Du Citation2018; Yang and Li Citation2018; DeWalt et al. Citation2020). Till now, only the complete mitochondrial genome of Acroneuria hainana Wu from China mainland in genus Acroneuria has been published (Huang et al. Citation2015). In this study, we sequenced and annotated the species of A. carolinensis which was from Canada and U.S.A. which represents the first record for continental North America to provide mitochondrial genome data for future studies of the order Plecoptera.
The specimen used in this study was collected and identified by Boris C. Kondratieff from Sulfur Spring Fork (Hicksville, VA) (N 37.2156, W −81.0929) on 18 May 2017. Voucher specimen with the no. VHL-0148 is now deposited in Henan Institute of Science and Technology (HIST, Xinxiang, China). We extracted the total DNA from the adult’s sternal muscle of A. carolinensis by QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s protocol. The sequences were assembled by BioEdit version 7.0.5.3 (Hall Citation1999). Protein-coding genes (PCGs) were aligned and analyzed with MEGA 6.0 for the base composition (Tamura et al. Citation2013). Phylogenetic analyses were performed using Bayesian inference (BI) methods with MrBayes 3.2.6 (Ronquist et al. Citation2012).
The complete mitogenome of A. carolinensis which was deposited in the GenBank under the accession number MN969989 is 15,718 bp in length and contains 37 encoding genes and one big non-coding region. In general, the nucleotide composition of the mitogenome obviously preferred A/T nucleotides with 34.5% of A, 30.1% of T, 22.4% of C, 13.0% of G, and 64.6% of A + T content through the data analysis. The A + T content of all PCGs tRNAs, ribosomal RNAs (rRNAs) and the control region was 62.7%, 67.8%, 68.6%, and 73.9%, respectively. The initiation codon of 10 PCGs shared with the normal start codon except ND2, ND5, and ND1 which used the exceptional GTG, GTG, and TTG as start codon. All PCGs stopped with the conservative terminal codon TAA or TAG, while only COII gene used the single T as the stop signal. With an exception for tRNASer(AGN), 21 tRNAs have the typical cloverleaf secondary structure.
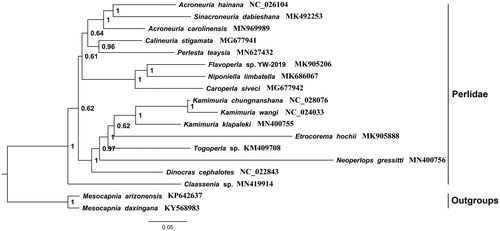
The phylogenetic analysis was based on the sequences of the PCG12R matrix (including the first and second codons of all PCGs and two rRNAs) from 16 species of Perlidae and two species of Capniidae (Mesocapnia arizonensis and Mesocapnia daxingana as outgroups) by using the BI method. As shown in , we found that the relationship in subfamily Acroneuriinae was ((((Acroneuria hainana + Sinacroneuria) + Acroneuria carolinensis) + (Calineuria + Perlesta)) + ((Flavoperla + Niponiella) +Caroperla)), and the genus Acroneuria and Sinacroneuria were clustered in one clade.
Figure 1. Phylogenetic analyses of A. carolinensis based on the PCG12R matrix (including PCGs first and second codons and two rRNAs) from 16 species of Perlidae and two species of Nemouridae by Bayesian inference (BI) method. The NCBI accession number for each species is indicated after the scientific name.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, accession number [MN969989], or available from the corresponding author.
Additional information
Funding
References
- Chen ZT, Du YZ. 2018. A checklist and adult key to the Chinese stonefly (Plecoptera) genera. Zootaxa. 4375(1):59–74.
- DeWalt RE, Maehr MD, Neu-Becker U, Stueber G. 2020. Plecoptera species file online; http://plecoptera.speciesfile.org/ [accessed 2020 Jun 15].
- Gutiérrez-Fonseca PE, Springer M. 2011. Description of the final instar nymphs of seven species from Anacroneuria Klapálek (Plecoptera: Perlidae) in Costa Rica, and first record for an additional genus in Central America. Zootaxa. 2965(1):16–38.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Huang MC, Wang YY, Liu XY, Li WH, Kang ZH, Wang K, Li XK, Yang D. 2015. The complete mitochondrial genome and its remarkable secondary structure for a stonefly Acroneuria hainana Wu (Insecta: Plecoptera, Perlidae). Gene. 557(1):52–60.
- Murányi D, Li WH. 2016. On the identity of some oriental Acroneuriinae taxa (Plecoptera: Perlidae), with an annotated checklist of the subfamily in the realm. Opusc Zool (Budapest). 47:173–196.
- Ronquist F, Teslenko M, van der Mark PVD, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Yang D, Li WH. 2018. Species catalogue of China. Vol. 2. Animals, Insecta (III), Plecoptera. Beijing: Science Press.
