Abstract
The first complete mitochondrial genome of the intertidal barnacle Fistulobalanus albicostatus Pilsbry, 1916 (Crustacea: Maxillopoda: Sessilia) is presented. The genome is a circular molecule of 15,665 bp, which encodes a set of 37 typical metazoan genes. All non-coding regions are 438 bp in length, with the longest one speculated as the control region (264 bp), which is located between srRNA and trnI. All protein-coding genes (PCGs) have an ATD (ATA, ATT, or ATG) start codon, except nad1, which is initiated with GTG. Remarkably, cox3, cob, nad1-5 have incomplete stop codons (T–– or TA–) and the remaining PCGs have the complete stop codon (TAA). Phylogenetic analysis based on 13 mitochondrial PCGs shows that the members of the Archaeobalanidae and Balanidae intermingle with species from Pyrgomatidae. The results supposed that Balanidae and Archaeobalanidae are non-monophyly.
Fistulobalanus albicostatus Pilsbry, 1916 (Crustacea: Maxillopoda: Sessilia) is a common acorn barnacle in mangroves and on soft shores, inhabiting on rocks, mollusk shells, tree trunks, and mangrove leaves (Prabowo and Yamaguchi Citation2005; Chang et al. Citation2017; Hayashi Citation2017). The species is widely distributed in East Asia, including Japan, Korea, and China (Newman and Ross Citation1976; Chan and Leung Citation2007; Ping-Hung et al. Citation2007). The work presents the first complete mitochondrial genome of F. albicostatus. Specimen of F. albicostatus was collected from Qidong (32°1′24″N, 121°44′21″E), Jiangsu Province, China. Genomic DNA was isolated using TIANamp Marine Animal DNA Kit (TIANGEN, Beijing, China) following the manufacturer’s protocol, which was stored at Marine Museum of Jiangsu Ocean University (accession number: Fal-002). The amplification of internal fragments and long fragments followed the procedures described in our previous study (Chen et al. Citation2019). PCR products were purified (GeneMark), cloned (pGEMT easy, Promega, Madison, WI) and sequenced (MAP BIOTECH, Shanghai, China).
The complete mitochondrial genome of F. albicostatus is a circular molecule of 15,665 bp in length, encodes 13 protein-coding genes (PCGs), 2 ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), and 1 non-coding region (GenBank accession number: MK617531). Four PCGs and two rRNAs are encoded on the light strand (nad1, nad4, nad4L, and nad5), while the other nine PCGs are located on the heavy strand.
The base composition was 36.0% A, 17.4% C, 12.1% G, and 34.5% T. The overall A + T content of the mitochondrial genome of F. albicostatus is 70.6%. Non-coding regions make up 438 bp, with the longest one speculated as the control region (264 bp), which is located between srRNA and trnI. The srRNA (AT content: 72.4%) and lrRNA (AT content: 74.6%) are 898 bp and 1302 bp, respectively. All PCGs have an ATD (ATA, ATT, or ATG) start codon, except nad1, which is initiated with GTG. Remarkably, cox3, cob, and nad1-5 have incomplete stop codons (T- or TA-) and the remaining PCGs have the complete stop codon (TAA).
To investigate the phylogenetic position and the inner relationships of the suborder Balanomorph, phylogenetic trees were constructed with nucleotide sequences of 13 PCGs from 22 complete mitochondrial genomes of Sessilia, and Lepas australis (NC_025295) as outgroup species, using maximum likelihood with MEGA7 (Kumar et al. Citation2016) (). In the tree, F. albicostatus is the most primitive member within the superfamily Balanoidea. Within the superfamily Balanoidea, the tree is divided into two parts. One part has two species of Megabalanus (M. ajax and M. volcano) cluster with Acasta sulcata, which belongs to Archaeobalanidae. Two are grouped with Balauns balanus, with being the most distantly related taxon. In another part, Nobia grandis and Savignium sp. BKKC-2013 are grouped together with high support (BP = 100), and then cluster with Armatobalanus allium and Striatobalanus amaryllis. The members of the Archaeobalanidae and Balanidae intermingle with species from Pyrgomatidae. The results supposed that Balanidae and Archaeobalanidae are non-monophyly, which is consistent with previous researches (Tsang et al. Citation2014, Citation2017; Shen et al. Citation2015).
Figure 1. Maximum-likelihood phylogenetic tree based on 13 PCGs nucleotide acid sequences of Fistulobalanus albicostatus and 22 other Cirripedia species. Nodal supports are denoted on the corresponding branches for a bootstrap value >50% for ML, while *represents the value ≤50%.
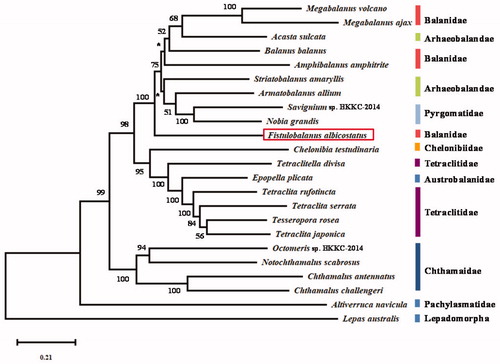
The accession numbers of the genomes used for comparison are NC_024636 (Megabalanus ajax), NC_006293 (Megabalanus volcano), NC_029168 (Acasta sulcata), NC_024525 (Amphibalanus amphitrite), NC_029167 (Armatobalanus allium), KJ_754821 (Savignium sp. BKKC-2014), NC_023945 (Nobia grandis), NC_029169 (Chelonibia testudinaria), NC_029170 (Tetraclitella divisa), NC_033393 (Epopella plicata), NC_037398 (Tetraclita rufotincta), NC_029154 (Tetraclita serrata), NC_037241 (Tesseropora rosea), NC_008974 (Tetraclita japonica), NC_026730 (Chthamalus antennatus), NC_022716 (Notochthamalus scabrosus), KJ_754820 (Octomeris sp. BKKC-2014), NC_005936 (Pollicipes polymerus), NC_025295 (Lepas australis), and NC_026576 (Lepas anserifera).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MK617531.
Additional information
Funding
References
- Chan BKK, Leung PTY. 2007. Antennular morphology of the cypris larvae of the mangrove barnacle Fistulobalanus albicostatus (Cirripedia: Thoracica: Balanomorpha). J Mar Biol Ass UK. 87(4):913–915.
- Chang YW, Chan JSM, Hayashi R, Shuto T, Tsang LM, Chu KH, Chan BKK. 2017. Genetic differentiation of the soft shore barnacle Fistulobalanus albicostatus (Cirripedia: Thoracica: Balanomorpha) in the West Pacific. Mar Ecol. 38(2):e12422.
- Chen P, Song J, Shen X, Cai Y, Chu KH, Li Y, Tian M. 2019. Mitochondrial genome of Chthamalus challengeri (Crustacea: Sessilia): gene order comparison within Chthamalidae and phylogenetic consideration within Balanomorpha. Acta Oceanol Sin. 38(6):25–31.
- Hayashi R. 2017. First documentation of the barnacle Fistulobalanus albicostatus (Cirripedia: Balanomorpha) as an epibiont of loggerhead sea turtle Caretta caretta. Mar Biodivers. 47(1):157–158.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Newman WA, Ross A. 1976. Revision of the balanomorph barnacles; including a catalog of species. Mem San Diego Soc Nat Hist. 9:1–108.
- Ping-Hung C, Yung-Hui C, I-Ming C. 2007. Effect of biofilm age and type on settlement of cyprids of the barnacle, Fistulobalanus albicostatus Pilsbry (Thoracica Balanidae). Zool Stud. 46:521–528.
- Prabowo RE, Yamaguchi T. 2005. A new mangrove barnacle of the genus Fistulobalanus (Cirripedia: Amphibalaninae) from Sumbawa Island. J Mar Biol Ass UK. 85(4):929–936.
- Shen X, Tsang LM, Chu KH, Achituv Y, Chan BK. 2015. Mitochondrial genome of the intertidal acorn barnacle Tetraclita serrata Darwin, 1854 (Crustacea: Sessilia): gene order comparison and phylogenetic consideration within Sessilia. Mar Genomics. 22:63–69.
- Tsang LM, Chu KH, Nozawa Y, Chan BK. 2014. Morphological and host specificity evolution in coral symbiont barnacles (Balanomorpha: Pyrgomatidae) inferred from a multi-locus phylogeny. Mol Phylogenet Evol. 77:11–22.
- Tsang LM, Shen X, Cheang CC, Chu KH, Chan BKK. 2017. Gene rearrangement and sequence analysis of mitogenomes suggest polyphyly of Archaeobalanid and Balanid barnacles (Cirripedia: Balanomorpha). Zool Scr. 46(6):729–739.
