Abstract
Musa acuminata var. chinensis is one of the most important wild banana species native to China which has huge potential breeding value by its cold tolerance and disease resistance. In this study, we first reported the complete chloroplast genome of M. acuminata var. chinensis and explore its phylogenetic position using a maximum likelihood phylogenetic tree. The chloroplast genome of M. acuminata var. chinensis is 170,402 bp in length and the overall GC content of the whole genome is 36.8%. It consisting of a pair of inverted repeat (IR, 35,320 bp) regions, a large single-copy (LSC, 88,870 bp) and a small single-copy (SSC, 10,900 bp). The chloroplast genome contained 112 genes, including 79 protein-coding genes, 29 tRNA genes, and 4 rRNA ribosomal genes. The most genes occur as a single copy, while 23 gene species occur in double copies. Phylogenetic analysis of 7 selected chloroplast genomes revealed that M. acuminata var. chinensis was closely related to M. acuminata ssp. malaccensis. The complete chloroplast genome of M. acuminata var. chinensis will greatly enhance precious gene resources for banana breeding programs in the future.
Musa acuminata Colla var. chinensis Häkkinen & Wang Hong belonging to Musa, Musacaea, is one of the most important wild banana species native to China which has huge potential breeding value due to its its cold tolerance and disease resistance. It is distributed northward along the China–Myanmar border and is found in river watersheds along these slopes, at altitudes between 300 and 800 m that tolerate slight frost damage. It also can grow under shaded tree cover. It also has basal, hermaphroditic female flowers and therefore self-pollination occurs before the floral bracts open (Häkkinen and Wang Citation2007). The information of chloroplast genomes has been extensively applied in understanding plant genetic diversity and evolution that are of great importance in breeding programs. The complete chloroplast genome of M. acuminata var. chinensis will greatly enhance precious gene resources for banana breeding programs in the future.
An individual of M. acuminata var. chinensis was collected from Yunnan, China (22°39.170′N, 103°03.763′E). Total genomic DNA was extracted from fresh young leaves using modified CTAB method (Doyle and Doyle Citation1987) and deposited at Hainan Tropical Ocean University (Accession number: M05). Purified total genomic DNA was used for sequencing. Whole-chloroplast genome sequencing was outsourced to Shanghai Majorbio Bio-pharm Technology Co., Ltd, on an Illumina Hiseq X Ten platform. Approximately 5.5 G clean reads were obtained and further assembled into contigs using MITObim v1.8 (Hahn et al. Citation2013) and SOAPdenovo v2.04 (Luo et al. Citation2012), with the reference genome of M.acuminata subsp. malaccensis (Genbank: HF677508.1) (Martin et al. Citation2013). The assembled chloroplast genome was mapped and annotated using the online program Organellar Genome Draw tool v1.2 (Lohse et al. Citation2013). The annotated chloroplast genomic sequence was submitted to Genbank under the accession number of MT593357.
The complete chloroplast genome of M. acuminata var. chinensis is 170,402 bp in length consisting of a pair of inverted repeat (IR, 35,320 bp) regions, a large single copy (LSC, 88,870 bp) and a small single copy (SSC, 10,900 bp). The chloroplast genome contained 112 genes, including 79 protein-coding genes, 29 tRNA genes, and 4 rRNA ribosomal genes. Most of the genes occur as a single copy, while 23 gene species occur in double copies, including 11 protein-coding genes (ndhA, ndhB, ndhH, rps7, rps12, rps15, rps19, rpl2, rpl23, ycf1, ycf2), 8 tRAN genes (trnI-CAU, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, trnN-GUU, trnL-CAA, trnH-GUG) and 4 rRNA genes (rrn16, rrn23, rrn4.5, rrm5). The overall GC content of the whole genome is 36.8% and the corresponding values in LSC, SSC and IR regions are 35.18%, 31.03% and 39.75%, respectively.
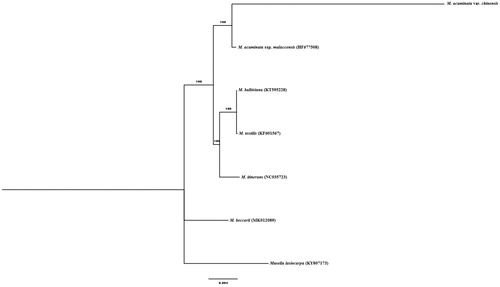
A total of six additional complete chloroplast genomes of the family Musaceae were used to clarify the phylogenetic position of M. acuminata var. chinensis. Musalla lasiocarpa (KY807173) (Zhang et al. Citation2018) was used as the outgroup. All of the chloroplast genomes were aligned in MAFFT (Katoh and Standley Citation2013) and a maximum likelihood tree was constructed in IQTREE v1.6 (Nguyen et al. Citation2015; Hoang et al. Citation2018) based on TVM + F + I with 1000 bootstrap replicates (). The phylogenetic tree reveals that M. acuminata var. chinensis was closely related to M. acuminata ssp. malaccensis compared to other species of Musa. The result provided valuable information for further banana breeding program.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in national center for biotechnology information (NCBI) at https://www.ncbi.nlm.nih.gov/, reference number [MT593357].
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Häkkinen M, Wang H. 2007. New species and variety of Musa (Musaceae) from Yunnan. China. Novon. 17(4):440–446.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short. Gigascience. 1(1):18.
- Martin G, Baurens F-C, Cardi C, Aury J-M, D'Hont A. 2013. The complete chloroplast genome of banana (Musa acuminata, Zingiberales): insight into plastid monocotyledon evolution . PLOs One. 8(6):e67350.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Zhang L, Guo X, Wang Z, Wang M, Hu Q. 2018. Characterization of the complete chloroplast genome of Musalla lasiocarpa. Mitochondrial DNA Part B. 3(2):728–729.