Abstract
We sequenced the complete mitochondrial genomes of Togoperla limbata which is the unique species distributed in Japan to provide supplementary data for future study in phylogenetic studies of Perlidae. This complete mitogenome of T. limbata is 15,915 bp long, including 37 typical genes and a control region as other stoneflies. Through the data analysis, the A + T content of the whole mitogenome, PCGs, tRNAs, rRNAs, and control region was 63.7%, 61.3%, 68.7%, 66.8%, and 74.8%. Most PCGs used typical start or stop codon except COI, ND5, and ND1 used the exceptional start codon and the COII and ND5 used the single T as the stop codon. Phylogenetic tree was constructed by using Bayesian inference (BI) and maximum-likelihood (ML) analysis based on the sequences of the 13 PCGs and two rRNAs and the result showed that subfamily Perlinae was a monophyletic group and the clade Togoperla and Paragnetina had a closer relationship.
Keywords:
There are 20 genera with more than valid 1100 species worldwide in family Perlidae, and a few genera extending into the southern hemisphere in Africa and South America (DeWalt et al. Citation2020). Genus Togoperla was originally established by Klapálek in Perlinae which includes about 13 valid species, and all of them are distributed in the Oriental region. Among them, eight species are distributed in China but only the species Togoperla limbata was found in Japan (Sivec et al. Citation1988; Stark and Sivec Citation2008; Chen and Du Citation2018; Yang and Li Citation2018; DeWalt et al. Citation2020). However, about nine mitochondrial genomes in seven genera of the Perlinae have previously been published but only one species in genus Togoperla (Qian et al. Citation2014; Wang et al. Citation2014; Elbrecht et al. Citation2015; Wang et al. Citation2016; Liu et al. Citation2019; Wei et al. Citation2020). Here, we sequenced the complete mitochondrial genomes of T. limbata which is the unique species distributed in Japan to provide supplementary data for future study in phylogenetic studies of Perlidae.
The specimen was collected by Murányi Dávid from Toon city, Ehime Pref, Japan (N33.4518, E132.5951) on 5 July 2015. Voucher specimen (no. VHL-0023) is now preserved in Henan Institute of Science and Technology (HIST), Henan Province, China at −20 °C. We extracted the total genomic DNA from adult’s chest muscle by using QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) and sequenced the sample by high-throughput sequencing method. The sequences were assembled by BioEdit version 7.0.5.3 (Hall Citation1999).
This complete mitogenome of T. limbata was deposited in the GenBank under the accession number MN969990 with 15,915 bp long, including 37 typical genes (13 protein-coding genes, 22 tRNA genes, and two rRNA genes) and a control region as other stoneflies. Through the data analysis, the A + T content of the whole mitogenome, PCGs, tRNAs, rRNAs, and control region biased toward A/T nucleotides was 63.7%, 61.3%, 68.7%, 66.8%, and 74.8%, respectively. Excluding the control region, the mitogenome has 10 intergenic regions, ranging from 1 to 20 bp. There are totally 51 bp overlapped nucleotides in 14 locations, ranging from 1 to 8 bp. With an exception for tRNASer(AGN) which lacks the dihydrouridine (DHU) arm, all tRNA genes can be folded into the normal cloverleaf secondary structure. Most PCGs used conservative initiation or terminal codon except COI, ND5, and ND1 used the exceptional start codon and the COII and ND5 used the single T as the stop codon.
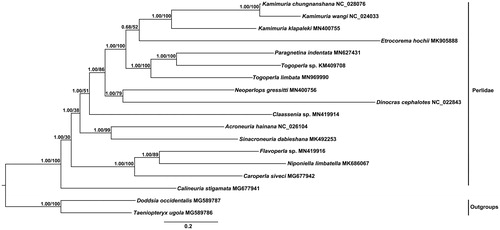
Phylogenetic tree was constructed by using Bayesian inference (BI) and maximum-likelihood (ML) analysis based on the sequences of the 13 PCGs and two rRNAs from 16 species in Perlidae and two species (Taeniopteryx ugola and Doddsia occidentalis as the outgroups) in Taeniopterygidae (). Two methods supported the identical tree topology, the subfamily Perlinae was a monophyletic group and the clade Togoperla and Paragnetina had a closer relationship. However, the monophyly of the genus Togoperla was not supported and more mitogenome information of Perlidae species is needed for further phylogenetic studies.
Figure 1. Phylogenetic trees of Togoperla limbata were inferred based on the PCGs of two rRNAs from 16 species in Perlidae and two species as the outgroups by Bayesian inference (BI) and maximum-likelihood (ML) methods. The NCBI accession number for each species is indicated after the scientific name.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, accession number [MN969990], or available from the corresponding author.
Additional information
Funding
References
- Chen ZT, Du YZ. 2018. A checklist and adult key to the Chinese stonefly (Plecoptera) genera. Zootaxa. 4375(1):59–74.
- DeWalt RE, Maehr MD, Neu-Becker U, Stueber G. 2020. Plecoptera species file online;http://plecoptera.speciesfile.org/ [accessed 2020 Jun 15].
- Elbrecht V, Poettker L, John U, Leese F. 2015. The complete mitochondrial genome of the stonefly Dinocras cephalotes (Plecoptera, Perlidae). Mitochondrial DNA A. 26(3):469–470.
- Liu ZZ, Wang Y, Li WH, Cao JJ. 2019. The complete mitochondrial genome of a stonefly species, Etrocorema hochii (Plecoptera: Pelidae). Mitochondrial DNA B Resour. 4(2):2690–2691.
- Qian YH, Wu HY, Ji XY, Yu WW, Du YZ. 2014. Mitochondrial genome of the stonefly Kamimuria wangi (Plecoptera: Perlidae) and phylogenetic position of Plecoptera based on mitogenomes. PLOS One. 9:e86328.
- Wang K, Ding SM, Yang D. 2016. The complete mitochondrial genome of a stonefly species, Kamimuria chungnanshana Wu, 1948 (Plecoptera: Perlidae). Mitochondrial DNA Part A. 27(5):3810–3811.
- Wang K, Wang YY, Yang D. 2014. The complete mitochondrial genome of a stonefly species, Togoperla sp. (Plecoptera: Perlidae). Mitochondrial DNA Part A. 27:1–1704.
- Wei X, Cao J, Wang P, Wang Y, Li W. 2020. The mitochondrial genome analysis of Paragnetina indentata (Plecoptera: Perlidae). Mitochondrial DNA B Resour. 5(1):44–45.
- Sivec I, Stark BP, Uchida S. 1988. Synopsis of the world genera of Perlinae (Plecoptera: Perlidae). Scopolia. 16:17.
- Stark BP, Sivec I. 2008. The genus Togoperla Klapálek (Plecoptera: Perlidae). Illiesia. 4:208–225.
- Yang D, Li WH. 2018. Species catalogue of China. Vol. 2. Animals, Insecta (III), Plecoptera. Beijing: Science Press.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
