Abstract
The complete chloroplast genome of Chrysosplenium ramosum Maxim. and Chrysosplenium alternifolium L. were reported in this study. The chloroplast genomes were 153,460 bp for C. ramosum and 152,619 bp for C. alternifolium. LSC and SSC of 83,670 bp and 17,342 bp were separated by two IRs of 26,224 bp each in C. ramosum. While C. alternifolium contained IRs of 25,992 bp, LSC of 83,524 bp and SSC of 17,111 bp. The chloroplast genome of C. ramosum contains 112 unique genes, including 79 protein-coding genes, four ribosomal RNA genes, and 30 transfer RNA genes. And the chloroplast genome of C. alternifolium contains 112 unique genes, including 79 protein-coding genes, four ribosomal RNA genes, and 30 transfer RNA genes. In addition, the rps12 gene was recognized as a trans-spliced gene and 17 intron-containing genes were also detected.
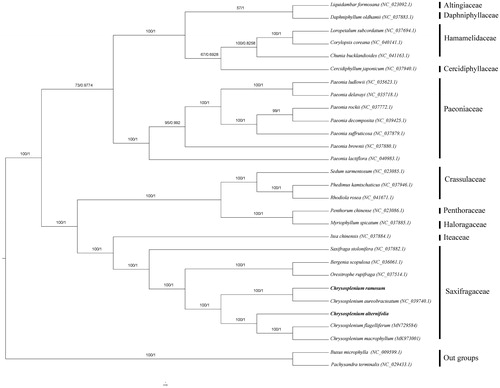
Chrysosplenium L. (Saxifragaceae) comprises about 65–70 species in worldwide, mainly distributed in the northern hemisphere (Hara Citation1957; Soltis et al. Citation2001; Lan et al. Citation2018; Liao et al. Citation2019). Due to the different characters: four sepals of flowers, lack petals, and have two equal or distinctly unequal fruiting capsules, Chrysosplenium is important in the taxonomy of Saxifragaceae (Maximowicz Citation1872). Chrysosplenium is divided into two subgenera: Alternifolia Franchet and Oppositifolia Franchet according to leaf arrangement (Hara Citation1957). In this study, we report and characterize two chloroplasts genomes from two species: Chrysosplenium ramosum from subgenus Oppositifolia and Chrysosplenium alternifolium from subgenus Alternifolia. Using these data, we reconstruct the phylogenetic tree of this genus to reveal the relationship and provide useful information for further study of Chrysosplenium.
The materials of C. ramosum and C. alternifolium were collected from Jilin, China (E 127° 30′ 30″, N 42° 10′ 27″) and Shimane-ken, Japan (E 132° 36′ 11″, N 35° 08′ 22″). The voucher specimens were deposited at the Herbarium of South-Central University for Nationalities (HSN) and the specimen accession numbers are SJH2017052107372 and RG2019032810003. The complete genomic DNA of C. ramosum and C. alternifolium were extracted using CTAB method and sequenced using the Illumina platform at Biomarker Technologies (Beijing, China). After filtered the low-quality data and adaptors, the obtained clean data were aligned to C. macrophyllum (MK973001) with bwa-0.7.12. The aligned reads were then assembled with ABYSS-2.0.2 (Jackman et al. Citation2017) after the best Kmer was chosen with kmergenie (Medvedev and Chikhi Citation2014). Then, connected the overlap and scaffolding again by SSPACE_Standard_v3.0. Finally, the gaps were filled by Sanger.
The complete chloroplast genomes of these two species exhibited a general quadripartite structure. C. ramosum (MK973002) was 153,460 bp in length and included a large single-copy (LSC) region of 83,670 bp and a small single-copy (SSC) region of 17,342 bp separated by two reverse repeated regions of 26,224 bp, the average GC content was 37.38%. On the other hand, the complete chloroplast genome size of C. alternifolium (MT362050) was 152,619 bp in length, the average GC content was 37.47%. Separating LSC of 83,524 bp and SSC of 17,111 bp, a pair of IRs was 25,992 bp long in each. The chloroplast genome of C. ramosum contains 130 genes, including 85 protein-coding genes, eight ribosomal RNA genes, and 37 transfer RNA genes. The chloroplast genome of C. alternifolium contains 130 genes, including 85 protein-coding genes, eight ribosomal RNA genes, and 37 transfer RNA genes.
Phylogenetic analysis was performed using whole chloroplast coding sequences of C. ramosum and C. alternifolium, combining with three species of Chrysosplenium, 22 species of Saxifrageles, and two species of Buxaceae as outgroup. The phylogenetic relationships were reconstructed by means of maximum-likelihood (ML) with the model of GTR + F+R3 and Bayesian inference analysis using the Markov Chain Monte Carlo method with 200,000 generations and sampling trees every 100 generations. Based on the phylogenetic tree, we can see that the five species of Chrysosplenium were clustered to a clade (). What’s more, the five species of Chrysosplenium seemed to divide into two clades by two subgenera.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in Genbank (https://www.ncbi.nlm.nih.gov/genbank/) according to their Genbank accession.
Additional information
Funding
References
- Hara H. 1957. Synopsis of the genus Chrysosplenium (Saxifragaceae). J Fac Sci Univ Tokyo. 7:1–90.
- Jackman SD, Vandervalk BP, Mohamadi H, Chu J, Yeo S, Hammond SA, Jahesh G, Khan H, Coombe L, Warren RL, et al. 2017. ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 27(5):768–777.
- Lan DQ, Liu H, Huang W, Yi LS, Zhang DD, Qin ED, Qin R. 2018. Investigation and Analysis of Chrysosplenium resources in Tibetan-inhabited area. JPGR. 20:662–668.
- Liao R, Dong X, Wu ZH, Qin R, Liu H. 2019. Complete chloroplast genome sequence of Chrysosplenium sinicum and Chrysosplenium lanuginosum (Saxifragaceae). Mitochondrial DNA B. 4(2):2142–2143.
- Maximowicz CJ. 1872. Diagnoses plantarum novarum Japoniae et Mandshuriae. St. Petersburg, sér. 3: BuLletin de l’Academie Imperiale des Sciences de Saint-Pétersbourg.
- Medvedev P, Chikhi R. 2014. Informed and automated k-mer size selection for genome assembly. Bioinformatics. 30(1):31–37.
- Soltis DE, Tago-Nakazawa M, Xiang QY, Kawano S, Murata J, Wakabayashi M, Hibsch-Jetter C. 2001. Phylogenetic relationships and evolution in Chrysosplenium (Saxifragaceae) based on matK sequence data. Am J Bot. 88(5):883–893.