Abstract
The melon fly, Zeugodacus cucurbitae (Diptera:Tephritidae) is an important invasive pest and distributed throughout tropical, subtropical countries and areas. In this study, we report the complete mitochondrial genome of the fly with 15, 685 base pair in length, which includes 37 genes (the large and small ribosomal RNA subunits, 22 transfer RNA genes, 13 genes encoding mitochondrial proteins) and a non-coding A + T-rich control region. Molecular phylogeny indicated that there was a high bootstrap value supported among Z. cucurbitae and Z. tau belonging to the Z. tau complex.
Melon fly, Zeugodacus cucurbitae (Diptera: Tephritidae), is an important invasive pest in fruits and vegetables (Matthew and Jang Citation2010), infesting primarily the fruit and flower of Cucurbitaceae (Vargas et al. Citation2015). Although native to India, it is now widely distributed in tropical, sub-tropical and temperate regions worldwide (Dhillon et al. Citation2005; Virgilio et al. Citation2010). In this study, we reported the complete mitogenome of Zeugodacus cucurbitae from Kunming, Southwest China. The result would be greatly helpful for phylogenetics, population genetics, and species identification.
The adult male flies were caught from Kunming (25.05°N, 102.76°E), Southwest China, on 12 July 2018. Specimen was deposited in the museum of Southwest Forestry University (Voucher ZCKM20180712), Kunming, China. Whole genomic DNA was extracted from adult fly following the manufacturer’s instruction in the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) and then sequenced and assembled using Illumina’s HiSeq2000 platform (Illumina, San Diego, CA). The sequence was preliminarily aligned within the CLUSTAL X program in BioEdit software. Protein-coding genes (PCGs), rRNA genes were predicted by using MITOS tools (Bernt et al. Citation2013), and tRNA was done through tRNAscan-SE (Lowe and Chan Citation2016).
The complete mitogenome of Z. cucurbitae is 15,685 bp in length (GenBank MH900082), containing 13 PCGs, 22 transfer RNA genes, 2 ribosomal RNA genes, and a major non-coding region known as the CR (control region). The base composition is 38.49% for A, 34.34% for T, 16.67% for C, and 10.50% for G. A total of 23 genes, including 9 PCGs(NAD2-3, NAD6, COX1-3, CYTB, ATP6, and ATP8) and 14 tRNAs, were distributed on the J-strand of mitogenome, while the remaining 14 genes, including 4 PCGs (NAD1, NAD4, NAD4l, and NAD5), 8 tRNAs, and 2 rRNAs (16S rRNA and 12S rRNA), are located on the N-strand. The genes arrangement is conservative and identical to the most common type of the ancestor insect Drosophila yakuba (Clary and Wolstenholme Citation1985).
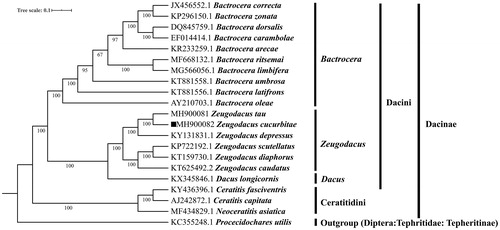
A phylogenetic tree was established based on complete mitogenome of Z. cucurbitae and other 20 species published Tephritidae species having Refseqs in NCBI by BI method in MrBayes version 3.2.6 (Huelsenbeck and Ronquist Citation2001; Dereeper et al. Citation2010). The result indicated that there was 100% bootstrap supported among Z. cucurbitae and Z. tau, which belong to Z. tau complex (). Furthermore, four clades were correctly identified as assigned Bactrocera, Zeugoacus, Dacus and Ceratitidini with high bootstrap confidence, while Zeugoacus and Dacus form a sister group (). In conclusion, the mitochondrial genome of Z. cucurbitae educed in the present study can provide essential DNA molecular data for further phylogenetic and evolutionary analysis and it will facilitate the development of new DNA markers for species diagnosis, therefore it will be helpful for improving accurate detection of quarantine species.
Figure 1. Molecular phylogeny for Zeugodacus cucurbitae and the related species in subfamily Dacinae based on complete mitogenome. Tree was constructed by Bayesian Inference (BI) method. Genbank accession numbers lie before the scientific name of species. The position of Z. cucurbitae is marked with solid square shape.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MH900082.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22(3):252–271.
- Dereeper A, Audic S, Claverie JM, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 10(1):8.
- Dhillon MK, Singh R, Naresh JS, Sharma HC. 2005. The melon fruit fly, Bactrocera cucurbitae: a review of its biology and management. J Insect Sci. 5:40–56.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: search and contextual analysis of transfer RNA genes. Nucl Acids Res. 44:54–57.
- Matthew SS, Jang EB. 2010. Cucumber volatile blend attractive to female melon fly, Bactrocera cucurbitae (Coquillett). J Chem Ecol. 36(7):699–708.
- Vargas RI, Piñero JC, Leblanc L. 2015. An overview of pest species of Bactrocera fruit Flies (Diptera: Tephritidae) and the integration of biopesticides with other biological approaches for their management with a focus on the pacific region. Insects. 6(2):297–318.
- Virgilio M, Delatte H, Backeljau T, DE Meyer M. 2010. Macrogeographic population structuring in the cosmopolitan agricultural pest Bactrocera cucurbitae (Diptera:Tephritidae). Mol Ecol. 19(13):2713–2724.
