Abstract
The complete mitogenome of Corythoxestis sunosei (GenBank accession number MT611524) is 15,511 bp in length, and harbors 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes, and an A + T-rich region. The overall base composition is A (40.04%), C (10.64%), G (7.62%), and T (41.70%), showing AT-rich feature (81.74%). ATG, ATT, CGA were initiation codons and TAA and T were termination codons. All the 22 tRNAs displayed a common cloverleaf secondary structure, except for trnS1 which lacked the dihydrouracil (DHU) arm. Phylogenetic tree based on 13 PCGs showed that C. sunosei has a close phylogenetic relationship with Gibbovalva kobusi and Cameraria ohridella and belongs to Gracillariidae.
The leaf-mining moth Corythoxestis sunosei belongs to the family Gracillariidae, which includes about 2556 described species in 151 genera (De Prins and De Prins Citation2019). C. sunosei was found for the first time mining the leaves of Chinese medical plant Uncaria rhynchophylla in China (Jiang et al. Citation2020). In this study, adult specimens of C. sunosei were collected from Jianhe County, Guizhou Province, China (N27°52′, E108°47′), and deposited in the insect specimen room of Shandong Normal University with an accession number SDNU. Ent005642–5653.
The complete mitogenome of C. sunosei (GenBank accession number MT611524) is 15,511 bp in length, and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (16S rRNA and 12S rRNA), and an A + T-rich region (putative control region) (Boore Citation1999). The gene order and organization of C. sunosei were identical to those observed in other moth mitogenomes (Peng et al. Citation2017; Chen et al. Citation2019). The overall base composition of C. sunosei mitogenome were A (40.04%), C (10.64%), G (7.62%), and T (41.70%), with A + T content of 81.74%. The AT-skew and GC-skew of this mitogenome were −0.020 and −0.166, respectively. Fourteen genes (trnQ, trnC, trnF, trnH, trnY, trnL1, trnP, trnV, nad1, nad4, nad4L, nad5,12S rRNA, and 16S rRNA) were encoded on the minority strand (N-strand) of the mitogenome, whereas the remaining genes were encoded on the majority strand (J-strand). Gene overlaps were present at 11 gene junctions and involved a total of 33 bp; the longest overlap (8 bp) is located between trnW and trnC. There are 10 intergenic spacer regions comprising a total of 121 bp with length varying from 1 to 56 bp. The largest intergenic spacer is located between nad2 and trnQ. The length of 22 tRNAs varied from 61 bp (trnC) to 71 bp (trnK), and A + T content varied from 71.21% (trnL2) to 92.42% (trnE). All the 22 tRNAs displayed a common cloverleaf secondary structure, except for trnS1 (AGN) which lacked the dihydrouracil (DHU) arm. The 16S rRNA is located between trnL1 and trnV, with length of 323 bp and A + T content of 86.47%. The 12S rRNA is located between trnV and control region, with length of 720 bp and A + T content of 85.14%. The control region is located between 12S rRNA and trnM is 735 bp in length with an A + T content of 94.97%.
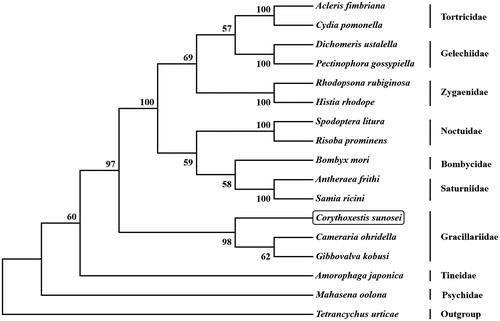
The initial codons for 12 PCGs were cononical putative start condons ATN (ATG for cox2, cox3, cob, atp6, nad1, nad4, nad4L, and nad6; ATT for atp8, nad2, nad3, and nad5), whereas cox1 use CGA as start codon. Ten PCGs terminate with the stop codon TAA; however, cob, nad4, and nad5 use incomplete T as stop codon, respectively. Based on the concatenated amino acid sequences of 13 PCGs, the neighbor-joining method was used to construct phylogenetic relationship of C. sunosei and 15 other moths with MEGA7 (Kumar et al. Citation2016). The result showed that C. sunosei has a close phylogenetic relationship with Gibbovalva kobusi and Cameraria ohridella (), which agree with the morphology taxonomy of the family Gracillariidae.
Figure 1. Phylogenetic tree showing the relationship between Corythoxestis sunosei and 15 other mothes based on neighbor-joining method. GenBank accession numbers used in this study are the following: Acleris fimbriana (HQ662522), Amorophaga japonica (MH823253), Antheraea frithi (NC_027071), Bombyx mori (NC_002355), Cameraria ohridella (KJ508042), Corythoxestis sunosei (MT611524), Cydia pomonella (JX407107), Dichomeris ustalella (KU366706), Gibbovalva kobusi (MK956103), Histia rhodope (MF542357), Mahasena oolona (NC_036410), Pectinophora gossypiella (KM225795), Rhodopsona rubiginosa (KM244668), Risoba prominens (KJ396197), Samia ricini (NC_017869), Spodoptera litura (KF701043), Tetrancychus urticae (EU345430). T. urticae was used as an outgroup. The moth determined in this study is boxed.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference number MT611524.
Additional information
Funding
References
- Boore JL. 1999. Survey and summary: animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Chen L, Liao CQ, Wang X, Tang SX. 2019. The complete mitochondrial genome of Gibbovalva kobusi (Lepidoptera: Gracillariidae). Mitochondrial DNA B. 4(2):2769–2770.
- De Prins J, De Prins W. 2019. Global taxonomic database of Gracillariidae (Lepidoptera). http://www.gracillariidae.net
- Jiang ZF, Zhang ZT, Liu HX, Liu TT. 2020. A new pest on Chinese traditional medical plant Uncaria rhynchophylla-Corythoxestis sunosei (Lepidoptera: Gracillariidae). J Environ Entomol. http://hjkcxb.alljournals.net/ch/reader/view_abstract.aspx?flag=2&file_no=201912060000001&journal_id=hjkcxb
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Peng SY, Jia WR, Huang ZJ, Wang YC, Li Y, Huang ZR, Zhang YF, Zhang X, Ding JH, Geng XX, et al. 2017. Complete mitochondrial genome of Histiarhodope Cramer (Lepidoptera: Zygaenidae. Mitochondrial DNA B. 2(2):636–637.
