Abstract
In this study, the complete mitochondrial genome of Clavulina sp. was sequenced and assembled. The complete mitochondrial genome of Clavulina sp. contains 20 protein-coding (PCG) genes, 2 ribosomal RNA (rRNA) genes, and 25 transfer RNA (tRNA) genes. The total size of the Clavulina sp. complete mitochondrial genome is 31,816 bp, with the GC content of 27.72%. Phylogenetic analysis indicated that the mitochondrial genome of Clavulina sp. exhibited a close relationship with that of the genus Cantharellus.
The genus Clavulina is an ectomycorrhizal fungal group distributed in many countries (Olariaga et al. Citation2009; Henkel et al. Citation2011). Most of species from the Clavulina genus are edible, and some species also show medicinal values (Khaund and Joshi Citation2014; Deo et al. Citation2019). Dozens of species have been described in genus Clavulina (Thacker and Henkel Citation2004; Uehling et al. Citation2012a, Citation2012b). Limited and varied morphological characteristics make it difficult to identify and classify Clavulina species accurately only by morphology (Thacker and Henkel Citation2004; Olariaga et al. Citation2009; Uehling et al. Citation2012a, Citation2012b). Mitochondrial genome is widely used to analyze the evolution and phylogeny of species (Li, He, et al. Citation2020; Wang et al. Citation2020; Li et al. Citation2020). However, up to now, no mitochondrial genome of Clavulina species has been published. The mitochondrial genome of Clavulina sp. will promote the understanding of the evolution, phylogeny, and taxonomy of this important ectomycorrhizal fungal group.
The specimen (Clavulina sp.) was collected from a mountain in Chuxiong, Yunnan, China (101.41 E; 25.12 N). The specimen was stored in Culture Collection Center of Chengdu University (No. Clasp08). The complete mitochondrial genome of Clavulina sp. was assembled according to previously described methods (Li, Xiang, et al. Citation2019; Wang et al. Citation2020). The total genomic DNA of Clavulina sp. was extracted using a Fungal DNA Kit D3390-00 (Omega Bio-Tek, Norcross, GA). A Gel Extraction Kit (Omega Bio-Tek, Norcross, GA) was used to purify the extracted total DNA. We stored the purified DNA in Chengdu University (No. DNA_ Clasp08). Sequencing libraries were constructed with the purified genomic DNA using a NEBNext® Ultra™ II DNA Library Prep Kit (NEB, Beijing, China). We conducted whole genomic sequencing (WGS) of Clavulina sp. using the Illumina HiSeq 2500 Platform (Illumina, SanDiego, CA). The mitochondrial genome of Clavulina sp. was de novo assembled using SPAdes version 3.9.0 (Bankevich et al. Citation2012). We obtained the complete mitogenome of Clavulina sp., and then annotated it according to the methods described by Li, Chen, et al. (Citation2018), Li, Ren, et al. (Citation2019), and Li, Wang, et al. (Citation2018).
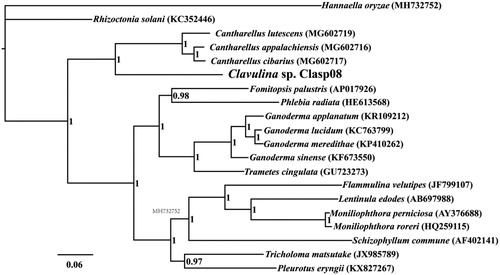
The complete mitochondrial genome of Clavulina sp. is 31,816 bp in length, with the base composition as follows: A (34.94%), T (37.32%), G (14.38%), and C (13.35%). The complete mitochondrial genome of Clavulina sp. contains 20 protein-coding genes, 2 ribosomal RNA genes (rns and rnl), and 25 transfer RNA (tRNA) genes. To investigate the phylogenetic positions of Clavulina sp., we constructed a phylogenetic tree for 20 species. The phylogenetic tree was constructed using the Bayesian analysis (BI) method based on the combined 14 core protein-coding genes according to methods described by Li, Wang, Jin, Chen, Xiong, Li, Liu, et al. (Citation2019), Li, Wang, Jin, Chen, Xiong, Li, Zhao, et al. (Citation2019) and Li, Yang, et al. (Citation2020). As shown in the phylogenetic tree (), the mitochondrial genome of Clavulina sp. exhibited a close relationship with that of the genus Cantharellus (Li, Liao, et al. Citation2018).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
This mitogenome of Clavulina sp. was submitted to GenBank under the accession number of MT649302 (https://www.ncbi.nlm.nih.gov/nuccore/ MT649302).
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Deo GS, Khatra J, Buttar S, Li WM, Tackaberry LE, Massicotte HB, Egger KN, Reimer K, Lee CH. 2019. Antiproliferative, immunostimulatory, and anti-inflammatory activities of extracts derived from mushrooms collected in Haida Gwaii, British Columbia (Canada). Int J Med Mushrooms. 21(7):629–643.
- Henkel TW, Aime MC, Uehling JK, Smith ME. 2011. New species and distribution records of Clavulina (Cantharellales, Basidiomycota) from the Guiana Shield. Mycologia. 103(4):883–894.
- Khaund P, Joshi SR. 2014. DNA barcoding of wild edible mushrooms consumed by the ethnic tribes of India. Gene. 550(1):123–130.
- Li Q, Chen C, Xiong C, Jin X, Chen Z, Huang W. 2018. Comparative mitogenomics reveals large-scale gene rearrangements in the mitochondrial genome of two Pleurotus species. Appl Microbiol Biotechnol. 102(14):6143–6153.
- Li Q, He X, Ren Y, Xiong C, Jin X, Peng L, Huang W. 2020. Comparative mitogenome analysis reveals mitochondrial genome differentiation in Ectomycorrhizal and asymbiotic Amanita species. Front Microbiol. 11:1382.
- Li Q, Liao M, Yang M, Xiong C, Jin X, Chen Z, Huang W. 2018. Characterization of the mitochondrial genomes of three species in the ectomycorrhizal genus Cantharellus and phylogeny of Agaricomycetes. Int J Biol Macromol. 118:756–769.
- Li Q, Ren Y, Shi X, Peng L, Zhao J, Song Y, Zhao G. 2019. Comparative mitochondrial genome analysis of two Ectomycorrhizal fungi (Rhizopogon) reveals dynamic changes of intron and phylogenetic relationships of the subphylum Agaricomycotina. Int J Mol Sci. 20(20):5167.
- Li Q, Wang Q, Chen C, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W. 2018. Characterization and comparative mitogenomic analysis of six newly sequenced mitochondrial genomes from ectomycorrhizal fungi (Russula) and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol. 119:792–802.
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Liu Q, Huang W. 2019. Characterization and comparative analysis of six complete mitochondrial genomes from ectomycorrhizal fungi of the Lactarius genus and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol. 121:249–260.
- Li Q, Wang Q, Jin X, Chen Z, Xiong C, Li P, Zhao J, Huang W. 2019. Characterization and comparison of the mitochondrial genomes from two Lyophyllum fungal species and insights into phylogeny of Agaricomycetes. Int J Biol Macromol. 121:364–372.
- Li Q, Xiang D, Wan Y, Wu Q, Wu X, Ma C, Song Y, Zhao G, Huang W. 2019. The complete mitochondrial genomes of five important medicinal Ganoderma species: features, evolution, and phylogeny. Int J Biol Macromol. 139:397–408.
- Li Q, Yang L, Xiang D, Wan Y, Wu Q, Huang W, Zhao G. 2020. The complete mitochondrial genomes of two model Ectomycorrhizal fungi (Laccaria): features, intron dynamics and phylogenetic implications. Int J Biol Macromol. 145:974–984.
- Olariaga I, Jugo BM, Garcia-Etxebarria K, Salcedo I. 2009. Species delimitation in the European species of Clavulina (Cantharellales, Basidiomycota) inferred from phylogenetic analyses of ITS region and morphological data. Mycol Res. 113(Pt 11):1261–1270.
- Thacker JR, Henkel TW. 2004. New species of Clavulina from Guyana. Mycologia. 96(3):650–657.
- Uehling JK, Henkel TW, Aime MC, Vilgalys R, Smith ME. 2012a. New species and distribution records for Clavulina (Cantharellales, Basidiomycota) from the Guiana Shield, with a key to the lowland neotropical taxa. Fungal Biol. 116(12):1263–1274.
- Uehling JK, Henkel TW, Aime MC, Vilgalys R, Smith ME. 2012b. New species of Clavulina (Cantharellales, Basidiomycota) with resupinate and effused basidiomata from the Guiana Shield. Mycologia. 104(2):547–556.
- Wang X, Song A, Wang F, Chen M, Li X, Li Q, Liu N. 2020. The 206 kbp mitochondrial genome of Phanerochaete carnosa reveals dynamics of introns, accumulation of repeat sequences and plasmid-derived genes. Int J Biol Macromol. 162:209–219.