Abstract
Nassarius graniferus is a marine gastropod species in family Nassariidae. In the present study, we firstly determined the complete mitochondrial genome of N. graniferus by next-generation sequencing. The mitochondrial sequence is a circular molecule of 16,417bp in length, with the typical structure of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs) and 2 ribosomal RNA genes. The GC% across the genome is 30.7%.
The gastropod family Nassariidae, a speciose group containing more than 600 extant species within the order Neogastropoda, is distributed worldwide from intertidal zone to a depth of about 1000 m (Cernohorsky Citation1972; Galindo et al. Citation2016). Taxonomy of nassariids is mainly based on morphological characters, often fail to get more credible results (Cernohorsky Citation1984). The nassariids are delicious and popular seafood in Asia, but some of species could accumulate algal toxins that cause paralytic shellfish poisoning (Lin et al. Citation1999; Choi et al. Citation2006). Therefore, accurate species identification is of great significance to solve the food safety problem of nassariids. Here, we present annotated mitochondrial genome of Nassarius graniferus, a small intertidal nassariid species which inhabits in coral and weedy sand of tropical Indo-Pacific, to provide an additional genetic resource for further study of species identification.
The individual of N. graniferus was collected from the Yongle Atoll in the Xisha Islands, Hainan Province, China (16°27′51″N, 111°44′17″E) and stored in 95% ethanol at the Specimen library of Shellfish Genetics and Breeding Laboratory, Ocean University of China (accession number: OUC-SGB-N-YL13). The total genomic DNA was extracted from the foot muscle, using the TIANamp Marine Animals DNA Kit (Tiangen Biotech, Beijing, China) according to manufacturer’s recommended protocol, then stored at −4 °C for short-term use. The DNA was sequenced on an Illumina HiSeq X sequencer using a PE150 protocol. Short-read DNA sequences were assembled using NOVOPlasty 3.7.2 (Dierckxsens et al. Citation2017). The mitogenome date were annotated using MITOS web servers (Bernt et al. Citation2013) (http://mitos.bioinf.uni-leipzig.de/index.py) and ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/), gene boundaries were compared manually with published Nassariidae mitochondrial genome to identify protein-coding (PCGs), ribosomal RNA (rRNAs) and transfer RNA (tRNAs) genes. Additionally, the ribosomal (rrnS and rrnL) genes were also identified by their similarity to gene sequence of Nassarius foveolatus (MH346209.1). The nucleotide compositions were computed using MEGA 6 (Tamura et al. Citation2013).
The mitochondrial genome of N. graniferus is 16,417 bp in length and was deposited in the Genebank (Accession number: MT374078). It encodes 35 metazoan mitochondrial genes, including 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes, consistent with other nassariid mitochondrial genomes. The rrnS and rrnL genes are 966 bp and 1392 bp in length, respectively. The overall nucleotide composition is as follows: 30.7% for A, 14.6% for C, 16.1% for G, and 38.6% for T, with a total A + T content of 69.3%. The GC skew value is 4.78%. Twelve of the 13 PCGs start with ATG, while ND4 start with ATA. Nine genes are terminated by TAA, four of which are terminated by TAG. All of the PCGs were encoded on the (+) strand.
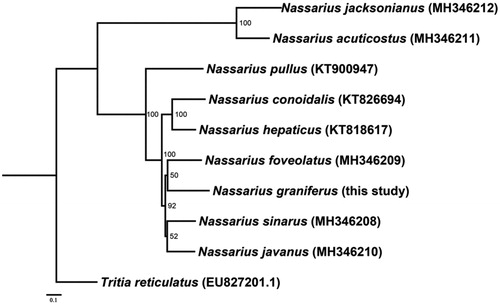
Phylogenetic relationships were inferred from available mitogenomes of 9 species of genus Nassarius using the maximum-likelihood (ML) method with 13 PCGs, and Tritia reticulatus was used as the outgroup. Maximum-Likelihood analysis was conducted using RAxML (Stamatakis Citation2014) and 1000 bootstraps were used to assess the support of nodes. The results of phylogenetic analysis showed that N. graniferus belongs to genus Nassarius (). This mitogenome sequence of N. graniferus would help in evolutionary biology, population genetics, and species diagnosis studies of Nassariinae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in National Center for Biotechnology Information at GenBank reference number MT374078 (https://www.ncbi.nlm.nih.gov/nuccore/MT374078).
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cernohorsky WO. 1972. Indo-Pacific Nassariidae (Mollusca: Gastropoda). Rec Auckl Inst Mus. 9:125–194.
- Cernohorsky WO. 1984. Systematics of the family Nassariidae (Mollusca: Gastropoda). Bull Auckl Inst Mus. 14:1–356.
- Choi M-C, Yu PKN, Hsieh DPH, Lam PKS. 2006. Trophic transfer of paralytic shellfish toxins from clams (Ruditapes philippinarum) to gastropods (Nassarius festivus). Chemosphere. 64(10):1642–1649.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18
- Galindo LA, Puillandre N, Utge J, Lozouet P, Bouchet P. 2016. The phylogeny and systematics of the Nassariidae revisited (Gastropoda, Buccinoidea). Mol Phylogenet Evol. 99:337–353.
- Lin Y, Jia X, Yang M, Zhong Y, Quan G, Fan P. 1999. Paralytic shellfish poison in contaminated shellfish along coast of China. Tropic Oceanol. 18:90–96.
- Stamatakis A. 2014. Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.