Abstract
Tagetes erecta L. is an important commercial and medicinal plant. In this study, we reported the complete chloroplast genome sequence of T. erecta. The genome has a circular structure of 152,076 bp containing a large single-copy region (LSC) of 83,914 bp, a small copy region (SSC) of 18,064 bp, and two inverted repeats (IR) of 25,049 bp by each. It harbors 111 unique genes, including 79 protein-coding genes, 4 ribosomal RNA genes, and 28 transfer RNA genes. A total of 41 microsatellite, 20 tandem, and 37 interspersed repeats were detected in the genome. The phylogenomic analysis shows that T. erecta is a single phylogenetic cluster. The complete chloroplast genome of T. erecta lays the foundation for the phylogenetic, evolutionary, and conservation studies of the genus Tagetes. Furthermore, the intergenic region of atpB-rbcL was variable among the species T. erecta. This suggests that this region might be a mutation hotspot and will be useful for phylogenetic study and the development of molecular markers. At last, we systematically identified the RNA editing sites in the chloroplast genome of T. erecta based on the transcriptome downloaded from the SRA database. This study identified the characteristics of the T. erecta chloroplast genome, SNPs, and RNA editing sites, which will facilitate species identification and phylogenetic analysis within T. erecta.
Introduction
Tagetes erecta L. belongs to the genus Tagetes of the family Asteraceae, as an annual ornamental plant and traditional Mexican medicine, is native to Mexico and South America. The genus Tagetes has 122 species (https://www.ipni.org/). Tagetes erecta is most well-known as an important commercial plant utilized mostly for the decorative purpose (Vasudevan et al. Citation1997; Ai et al. Citation2016; Ai et al. Citation2017) whose flower color can range from white to dark orange. Plants belonging to this genus have important medicinal value. Several studies have suggested that T. erecta has the potential to treat ailments, such as diabetes mellitus (Mudumbi et al. Citation2019). In particular, the flowers have been used to cure eye diseases, colds, conjunctivitis, coughs, ulcers, bleeding piles, and to purify the blood (Hemali and Sumitra Citation2014). Besides, Tagetes minuta L. is used as a medicinal tea in South America (Soule Citation1993). Illustration of the taxonomy classification and the development of efficient species discrimination markers of Tagetes species are fundamental for the development of medicinal products.
Many species of Tagetes were identified and reclassified based on morphological characteristics (Turner Citation1988; Schiavinato and Bartoli Citation2018). Unfortunately, many of the taxonomic classifications remain unresolved. For instance, studies have reinstated Tagetes pauciloba as a distinct species, which was previously treated as a synonymy of Tagetes filifolia (Schiavinato and Bartoli Citation2018). Morphological identification has significant limitations due to its strong dependence on the professional level and experience.
The DNA barcoding method can make up for the shortcomings of traditional methods because it is not affected by the environment, morphological, and sampling organs. The previous study has analyzed the phylogenetic relationship within the Tageteae based on the nuclear ribosomal ITS and chloroplast ndhF gene sequences, respectively (Loockerman et al. Citation2003). However, the trees from the two molecular makers were not completely congruent. The complete chloroplast genome sequence has more genetic information than the molecular marker sequence; it is widely used in angiosperm phylogenetic studies (Li et al. Citation2019). Consequently, the chloroplast genome sequence of T. erecta will promote the phylogenetic study and marker development of the tribe Tageteae.
Although the chloroplast genome sequence of T. erecta becomes publicly available, the intraspecific diversity of T. erecta is unknown. Different T. erecta lines might have various profiles of chemical compounds, and thus various biological activities. The medicinal products derived from different lines of T. erecta might have multiple efficacy and safety profiles. As a result, understanding the intraspecific diversity of T. erecta will be critical to ensure the consistent efficacy and safety profiles of the corresponding medicinal products. Furthermore, RNA-seq experiments have been performed with the leaf and flower tissues of T. erecta, which provided us an opportunity to characterization the RNA-editing events in the chloroplast of T. erecta.
Material and methods
Plant material, DNA extraction, and sequencing
The fresh leaves were collected from the Central China Medicinal Botanical Garden, EnShi, China (Geospatial coordinates: N30.177764, E109.743937). Genomic DNA was extracted with plant genomic DNA kit (Tiangen Biotech, China) and sequenced using the Hiseq 2500 platform (Illumina, San Diego, CA).
Genome assembly and annotation
The chloroplast genome was assembled from the raw sequence data by using NOVOPlasty (v.2.7.2) with the seed sequence of rbcL from Arabidopsis thaliana (Dierckxsens et al. Citation2017). The correctness of the assembly was validated by mapping all raw reads to the assembly using Bowtie 2 (v.2.0.1) (Langmead et al. Citation2009) under the default settings. The annotation of the chloroplast genome was conducted initially using CpGAVAS2 (Shi et al. Citation2019) and then edited using Apollo (Misra and Harris Citation2006). The genome sequence and annotations have been deposited in the GenBank with accession number MN309813.
Characteristics and repeat analysis
The codon usage and repeat analysis were analyzed using CpGAVAS2. The microsatellite sequence was analyzed with MISA software (Beier et al. Citation2017). The cutoff for the numbers of units for mono-, di-, tri-, tetra-, Penta-, and hexanucleotides were 10, 6, 5, 5, 5, and 5, respectively. The tandem repeats were analyzed by using TRF software (Benson Citation1999) with the size of repeat unit ≧ 7. The interspersed repeats were analyzed with VMATCH software (Kurtz et al. Citation2001). Both GC contents and codon usage were calculated using the program Cusp from EMBOSS (v6.3.1) (Langmead et al. Citation2009).
Phylogenetic analysis
The chloroplast genome sequence of T. erecta was compared against the sequences in the PlasDB database (http://www.herbalgenomics.org/plasdb). The whole chloroplast genome sequences of T. erecta and other 10 closely related species were used for phylogenetic analysis. The plastome gene sequences of 10 species were retrieved using the “DownloadCOG” module in PLasDB (http://www.herbalgenomics.org/plasdb). A total of 43 coding sequences (atpA, atpB, atpE, atpH, ndhA, ndhC, ndhD, ndhE, ndhG, ndhH, ndhJ, ndhK, petA, petG, petL, psaA, psaB, psaC, psaI, psaJ, psbA, psbB, psbC, psbD, psbF, psbH, psbI, psbJ, psbK, psbM, psbN, psbT, psbZ, rbcL, rpl20, rpl22, rps11, rps18, rps19, rps2, rps4, rps7, rps8) present in all of the 11 species were obtained. For the phylogenetic analysis, these protein sequences were aligned using the CLUSTALW2 (v2.0.12) program. The IQ-TREE2 (http://www.iqtree.org/) (Minh et al. Citation2020) was used to infer the evolutionary history, using the model of TVM + I. The bootstrap analysis was performed with 1000 replicates using UFBoot, Ultrafast Bootstrap Approximation (Minh et al. Citation2013).
Snp discovery in T. erecta chloroplast genome
During this study, another study published the chloroplast genome of T. erecta (NC_045211.1). To discover SNP between the two sequences of T. erecta chloroplast genome, we marked the sequence of NC_045211.1 as a reference and assembled them with the seqman module from DNASTAR Lasergene (v9). The SNP pipeline of seqman was used to identify SNPs with the default parameter.
Identification of RNA editing sites in T. erecta chloroplast genome
The RNA-Seq data from the flower and leaf (SRR6667676, SRR6667681) of T. erecta were downloaded from the GenBank SRA database (http://www.be-md.ncbi.nlm.nih.gov/sra). The cleaned reads from the two tissues were mapped to the chloroplast genome by bowtie2 (version 2.2.1) with mismatch = 7. RNA editing sites were called by REDItools (Picardi et al. Citation2015) with the following cutoffs: coverage ≥5, frequency ≥ 0.1, and p-value ≤ 0.05.
Results
General features of the chloroplast genome
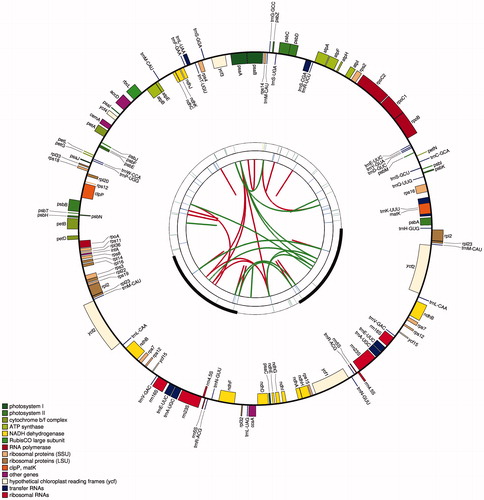
The chloroplast genome of T. erecta is 152,076 bp in size with a large single-copy region (LSC) of 83,914 bp, a small copy region (SSC) of 18,064 bp and two inverted repeats (IRs) of 25,049 bp by each (). There are 111 unique genes predicted in the chloroplast genome, including 79 protein-coding genes, 4 ribosomal RNA (rRNA) genes, and 28 transfer RNA (tRNA) genes (Table S1). Among these genes, 9 genes (rps16, rpoC1, atpF, petB, petD, rpl16, rpl2, ndhB, ndhA) contain only one intron, 2 genes (ycf3, clpP) contain two introns, and 6 tRNA genes (trnK-UUU, trnS-CGA, trnL-UAA, trnC-ACA, trnE-UUC, trnA-UGC) contain one intron (Table S2). The length of the protein-coding sequence (CDS) in the chloroplast genome of T. erecta is 71951 bp, representing 47.31% of the total genome length. In contrast, the length of the rRNA genes is 9050 bp, representing 5.95% of the total genome length, and the length of the tRNA genes is 2648 bp, representing 1.74% genome length.
Figure 1. The chloroplast genome of T. erecta created by using CPGAVAS2. The map contains four rings. From the center going outward, the first circle shows the scattered forward and reverse repeats connected with red and green arcs. The next circle shows the tandem repeats marked with short bars. The third circle shows the microsatellite sequences identified. The fourth circle shows the gene structure on the plastome. The genes were colored based on their functional categories, which are shown at the left corner.
The GC content analysis showed that the overall GC content is 37.38%, whereas that for the tRNA genes is 53.11%, that for the rRNA genes is 54.69%, and that for the protein-coding regions is 37.82%. We analyze the GC contents for the first, second, and third codon positions with the protein-coding regions. The GC contents of the third codon position are 29.85%, showed a higher AT representation. The GC contents of the three regions are ranked as IRs, LSC, and SSC, respectively. Moreover, a total of 24,321 codons were identified in the chloroplast genome of T. erecta. These include 64 unique codons for 20 amino acids and three termination codons. Among these codons, 2597 codons encode leucine, and 270 codes encode cysteine, respectively, representing the most and least abundant amino acids coded in the T. erecta chloroplast genome (Table S3).
Repeat analysis
Repeat sequences play an important role in genome evolution, such as insertion, deletion, rearrangement of large DNA segments, and can affect the length of the genome as well as the order of the genes (Tangphatsornruang et al. Citation2010). Here, we analyzed three kinds of repeat sequences (microsatellite repeats, tandem repeats, and interspersed repeats) in the chloroplast genome. For the microsatellite repeats, 41 (40 A/T and 1 AT/AT) were identified (Table S4). Only one compound microsatellite was identified, which is defined as two individual microsatellite repeats disrupted by less than 100 bases. Fewer microsatellites were found in the protein-coding regions than in the non-coding regions. The locations were further classified as intergenic spacers (IGS), exon, and intron. And numbers of microsatellites falling into these regions are 26, 10, and 5, respectively.
For the tandem repeats, 20 repeats were found in the chloroplast genome of T. erecta, meeting the two conditions that the length of the repeat unit is more than 30 bp and the similarity among the repeat unit sequences is more than 90% (Table S5). Most repeats have only two repeat units. And the lengths of repeat units range from 15 bp to 32 bp. For interspersed repeats, 17 palindromic repeats and 20 direct repeats were identified (Table S6). The most extended interspersed repeat unit is 49 bp long, and the two repeat units are located in the intron of the ycf3 gene and the intron of the ndhA gene, respectively. Whether or not this long tandem repeat played any role in the evolution of the chloroplast genome will be an interesting subject for future study.
Phylogenetic analysis
To examine the phylogenetic position of T. erecta, we analyzed the phylogeny between T. erecta and other 10 closely related species by IQ-TREE2 based on the protein-coding sequences shared in all the eleven chloroplast genomes (). In the PlasDB database, the 10 species closest to the T. erecta were selected for phylogenetic analysis which includes Guizotia abyssinica, Mikania micrantha, Galinsoga quadriradiata, Eclipta prostrata, Sphagneticola calendulacea, Eclipta alba, Ambrosia artemisiifolia, Parthenium argentatum, Helianthus hirsutus, and Helianthus strumosus, all of which belong to the family Asteraceae, subfamily Asteroideae. Eclipta alba was selected as the outgroup. The phylogenetic analysis showed T. erecta was a single phylogenetic cluster. This is consistent with what is expected because no chloroplast genome sequences belonging to the other species of Tagetes are available.
Figure 2. The phylogenetic tree of T. erecta and its closest relatives. Complete chloroplast genome sequences from Guizotia abyssinica, Mikania micrantha, Galinsoga quadriradiata, Eclipta prostrata, Sphagneticola calendulacea, Eclipta alba, Ambrosia artemisiifolia, Parthenium argentatum, Helianthus hirsutus, Helianthus strumosus were used to construct the tree using IQ-TREE.

Snp identification from the chloroplast genome
To discover SNPs from the chloroplast genome, we compared the two chloroplast genome of T. erecta and identified 139 SNPs, as shown in Table S7. Among them, 136 SNPs located in the intergenic region between the atpB and rbcL gene, and three SNPs located in the rbcL gene. The intergenic region between atpB and rbcL gene is hypervariable. The molecular markers based on the intergenic region between the atpB and rbcL gene might be effective in distinguishing T. erecta under the species taxa level.
Identification of RNA editing sites in T. erecta chloroplast genome
Plant organelle RNA editing is a post-transcriptional change in the nucleotide composition of an RNA (Freyer et al. Citation1997). To obtain the picture of RNA-editing in the chloroplast genome of T. erecta, we have investigated the occurrence of editing sites based on the transcriptome data of flower and leaf using REDItools. All RNA editing sites found in each tissue were shown in Table S8. There are nine RNA editing sites found across two tissues of flower and leaf. Two and nine unique RNA editing sites were found in flower and leaf tissues. The majority of editing events in the chloroplast genome of T. erecta are C-to-U transitions. The percentages of C-to-U edited sites were 90.9% and 88.9% in flower and leaf tissues, respectively. In the flower tissue, there were 10 RNA editing sites on the coding sequence of 8 genes, including atpI, psbZ, rps14, rbcL, accD, rpoA, and rpl23 ndhB. Only one RNA editing site was found in the intergenic region. In the leaf tissue, there were 15 RNA editing sites in the coding sequence of 11 genes, including rps2, psbZ, rps14, ndhJ, rbcL, accD, petL, petB, rpoA, rpl23 and ndhB. Three RNA editing sites were found in the intergenic regions.
Discussion
In this study, we sequenced and analyzed the complete chloroplast genome of T. erecta. Taking together with publicly available data, we carried out an intraspecific genetic variation study. Our phylogenetic study suggested that T. erecta was a single phylogenetic cluster. This is consistent with the classification based on the morphological characters. The data here are rather limited in its usefulness to resolve any problems in the taxonomic classification of Tagetes due to the limited sampling. This results from the difficulty in collecting the samples and carried out the reliable classification of these species. Nevertheless, those would be the focus of future research.
Comparing two T. erecta chloroplast genome sequences identified a total of 139 SNPs, which corresponds to an average of 0.9 SNP per kb sequences. Few studies have reported the nucleotide diversity among plastome sequences. For example, in one study, plastome sequences from four Panax ginseng lines were compared. The plastome sequences of three lines were identical, and the fourth one had a 1-bp insertion at base 5472 (Zhao et al. Citation2014). As a result, the level of nucleotide diversity in the genus Tagetes is higher than those in the genera Panax. One interesting aspect is that these SNPs are enriched in a particular locus. 136 of 139 SNPs (97.84%) were found in the intergenic regions between atpB and rbcL. The enrichment of nucleotide diversity in a particular region has not been reported before. Understand the underlying mechanism will be an interesting subject for future studies. Nevertheless, this region can be exploited for the development of high-resolution markers for intraspecific discrimination.
Mapping of the RNA-seq data to the references identified a total of 20 RNA-editing sites. In flowering plants, 30–40 such alterations are usually found in plastids (Takenaka et al. Citation2013). As a result, the occurrence of RNA-editing sites in T. erecta plastid is lower than in other species. The underlying mechanism will be an interesting subject for future studies.
In conclusion, the identification and characterization of the complete chloroplast genome sequence of T. erecta will help us identify Tagetes species with higher resolution and understand the relationship of related Tagetes species and to dissect the evolutionary history of Tagetes species. With the chloroplast genome of T. erecta available, sequencing and assembly of additional chloroplast genomes from varieties of T. erecta and other Tagetes species will become straightforward.
Author contributions
HMC conceived the study; MJ collected samples of T. erecta, extracted DNA for next-generation sequencing, assembled and validated the genome; YCX performed data analysis and drafted the manuscript; HMC, LQW, JTL and JY reviewed the manuscript critically. All authors have read and agreed on the contents of the manuscript.
tmdn_a_1791001_sm0348.zip
Download Zip (37.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/nuccore/1829069059.
Additional information
Funding
References
- Ai Y, Zhang C, Sun Y, Wang W, He Y, Bao M. 2017. Characterization and functional analysis of five MADS-Box B class genes related to floral organ identification in Tagetes erecta. PLOS One. 12(1):e0169777.
- Ai Y, Zhang Q, Wang W, Zhang C, Cao Z, Bao M, He Y. 2016. transcriptomic analysis of differentially expressed genes during flower organ development in genetic male sterile and male fertile Tagetes erecta by digital gene-expression profiling. PLOS One. 11(3):e0150892.
- Beier S, Thiel T, Munch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585.
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Freyer R, Kiefer-Meyer MC, Kossel H. 1997. Occurrence of plastid RNA editing in all major lineages of land plants. Proc Natl Acad Sci USA. 94(12):6285–6290.
- Hemali P, Sumitra C. 2014. Evaluation of antioxidant efficacy of different fractions of Tagetes erecta L. Flowers. IOSRJPBS. 9(5):28–37.
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29(22):4633–4642.
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25.
- Li HT, Yi TS, Gao LM, Ma PF, Zhang T, Yang JB, Gitzendanner MA, Fritsch PW, Cai J, Luo Y, et al. 2019. Origin of angiosperms and the puzzle of the Jurassic gap. Nat Plants. 5(5):461–470.
- Loockerman DJ, Turner BL, Jansen RK. 2003. Phylogenetic relationships within the Tageteae (Asteraceae) based on nuclear ribosomal ITS and chloroplast ndhF gene sequences. Syst Bot. 28:191–207.
- Minh BQ, Nguyen MA, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Misra S, Harris N. 2006. Using Apollo to browse and edit genome annotations. Curr Protoc. Bioinformatics Chapter. 9:Unit 9 :5.
- Mudumbi JBN, Daso AP, Okonkwo OJ, Ntwampe SKO, Matsha TE, Mekuto L, Itoba-Tombo EF, Adetunji AT, Sibali LL. 2019. Propensity of Tagetes erecta L., a medicinal plant commonly used in diabetes management, to accumulate perfluoroalkyl substances. Toxics. 7(1):18.
- Picardi E, D'Erchia AM, Montalvo A, Pesole G. 2015. Using REDItools to detect RNA editing events in NGS datasets. Curr Protoc Bioinformatics. 49:12.
- Schiavinato DJ, Bartoli A. 2018. About the identity of Tagetes pauciloba (Asteraceae, Tageteae). Phytotaxa. 362(2):200–210.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Soule J. 1993. Tagetes minuta: a potential new herb from South America. p. 649-654. In: J. Janick and J.E. Simon (eds.), New crops. Wiley, New York.
- Takenaka M, Zehrmann A, Verbitskiy D, Hartel B, Brennicke A. 2013. RNA editing in plants and its evolution. Annu Rev Genet. 47:335–352.
- Tangphatsornruang S, Sangsrakru D, Chanprasert J, Uthaipaisanwong P, Yoocha T, Jomchai N, Tragoonrung S. 2010. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res. 17(1):11–22.
- Turner BL. 1988. Two new species of Tagetes (Asteraceae-Tageteae) from Mexico. Phytologia. 65:129–131.
- Vasudevan P, Kashyap S, Sharma S. 1997. Tagetes: a multipurpose plant. Bioresour Technol. 62(1-2):29–35.
- Zhao Y, Yin J, Guo H, Zhang Y, Xiao W, Sun C, Wu J, Qu X, Yu J, Wang X, et al. 2014. The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Front Plant Sci. 5:696.
