Abstract
Acer sutchuenense subsp. tienchuanenge (Sapindaceae: Acer) is an endangered deciduous arbor species and endemic to China. Being obtained by using genome Illumina pair-end sequencing data, the complete chloroplast genome of A. sutchuenense subsp. tienchuanenge had a typical quadripartite structure, with 156,063 bp long, including a large single-copy (LSC) region of 85,772 bp, a small single-copy (SSC) region of 18,117 bp, and a pair of inverted repeats (IRs) (each 26,087 bp in length). A total of 136 genes were annotated, of which 113 are unique genes, including 30 tRNAs, 4 rRNAs, and 79 protein-coding genes. The overall GC content was 37.9%. The phylogenetic analysis suggested that A. sutchuenense subsp. tienchuanenge was the most closely related to A. griseum and A. triflorum. The complete chloroplast genome of A. sutchuenense subsp. tienchuanenge is valuable for assessment and conservation of genetic resources and further for phylogenetic study of Acer L.
Acer sutchuenense has two subspecies, which are A. sutchuenense subsp. sutchuenense and A. sutchuenense subsp. tienchuanenge. Acer sutchuenense subsp. tienchuanenge is a deciduous arbor species, mainly distributed in west and southwest of Sichuan province, while A. sutchuenense subsp. sutchuenense is distributed in Chongqing municipality, west of Hubei province and southwest of Hunan province (Wu et al. Citation2008). Except for the difference of geographical distribution, the trunk and leaves of A. sutchuenense subsp. tienchuanenge are larger than those of A. sutchuenense subsp. sutchuenense. According to the China Species Red List, A. sutchuenense was Critically Endangered (EN) (Wang and Xie Citation2004; Gibbs and Chen Citation2009). Till now only a few cases of study on investigation of wild resources and taxonomic status of A. sutchuenense (Zhang et al. Citation2011; Jiang et al. Citation2017; Wei Citation2019). Here, the complete chloroplast genome sequence of A. sutchuenense subsp. tienchuanenge was reported to provide a genomic resource and to clarify phylogenetic relationship of this plant with the other species in the Sapindaceae family.
Fresh leaves of A. sutchuenense subsp. tienchuanenge were collected from a wild individual tree from the Mount Emei, Sichuan province, China (N29°32′55′′, E103°21′28′′). The voucher specimen was deposited in Laboratory of Forest Silviculture and Tree Cultivation, Research Institute of Forestry, Chinese Academy of Forestry in Beijing, China (Voucher specimen: ACSUTCH-SCEM2019-01). The plant genomic DNA extraction kit (DP350) (Tiangen biotech Inc., Beijing, China) was used to extract the total genomic DNA, which was sequenced using the Illumina Hiseq platform (Huitong biotechnology Inc., Shenzhen, China). SPAdes version 3.9.0 and DOGMA were used for de novo assembly and annotation of the chloroplast genome (Wyman et al. Citation2004; Bankevich et al. Citation2012). The chloroplast genome map was drawn by using OGDRAW (Lohse et al. Citation2013).
The whole chloroplast genome of A. sutchuenense subsp. tienchuanenge had a typical quadripartite structure, with 156,063 bp long, including a large single-copy (LSC) region of 85,772 bp, a small single-copy (SSC) region of 18,117 bp, and a pair of inverted repeats (IRs) (each 26,087 bp in length). The total GC content is 37.9%. A total of 136 genes were annotated, of which 113 are unique genes, including 30 tRNAs, 4 rRNAs, and 79 protein-coding genes. The complete cp genome characteristics of A. sutchuenense subsp. tienchuanenge are similar to that of A. griseum (Wang et al. Citation2017).
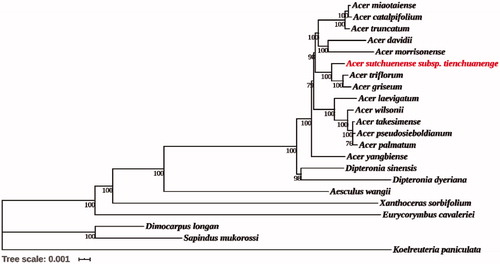
Twenty one complete chloroplast genomes were downloaded from NCBI Organelle Genome Database in order to reveal the phylogenetic position of A. sutchuenense subsp. tienchuanenge. Based on the GTR model, maximum-likelihood analysis and reconstruction of the phylogenetic relationship of 22 species were performed by MEGA version 7.0.14 (Kumar et al. Citation2016) (). All sequences were aligned using MAFFT (Nakamura et al. Citation2018). The results showed that A. griseum and A. triflorum were most closely related, followed by A. sutchuenense subsp. tienchuanenge, which was consistent with the morphological taxonomy, but inconsistent with the results of Wei (Citation2019), which indicated that A. sutchuenense and A. triflorum were the most closely related. In addition, 14 Acer species were clustered into a monophyly by 100% bootstrap value and have the closest relationship with Dipteronia, consistent with the result of Xia et al. (Citation2020). In short, the obtained chloroplast genome information of A. sutchuenense subsp. tienchuanenge was useful for the phylogenetic study of Acer L.
Figure 1. Phylogenetic tree reconstruction of 22 species using maximum likelihood (ML) based on the complete chloroplast genome sequences. There are the bootstrap support values from 1000 replicates given at each node. Their accession numbers are as follows: Acer catalpifolium: NC_041080; Acer davidii: NC_030331; Acer griseum: NC_034346; Acer pseudosieboldianum: NC_046487; Acer laevigatum: NC_042443; Acer miaotaiense: NC_030343; Acer morrisonense: NC_029371; Acer palmatum: NC_034932; Acer takesimense: NC_046488; Acer triflorum: NC_047296; Acer truncatum: NC_037211; Acer wilsonii: NC_040988; Acer yangbiense: MK479229; Aesculus wangii: NC_035955; Dimocarpus longan: NC_037447; Dipteronia dyeriana: NC_031899; Dipteronia sinensis: NC_029338; Eurycorymbus cavaleriei: NC_037443; Koelreuteria paniculata: NC_037176; Sapindus mukorossi: NC_025554; Xanthoceras sorbifolium: NC_037448.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, reference number MT216764.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Gibbs D, Chen YS. 2009. The red list of maples. Richmond (VA): Botanic Gardens Conservation International.
- Jiang ZG, Wang WH, Zhang JB. 2017. Study on rare and Endangered plants in Shennongjia. Hubei Agric Sci. 56(19):3651–3656.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Nakamura T, Yamada KD, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 34(14):2490–2492.
- Wang WC, Chen SY, Zhang XZ. 2017. The complete chloroplast genome of the Endangered Chinese paperbark maple, Acer griseum (Sapindaceae). Conservation Genet Resour. 9(4):527–529.
- Wang S, Xie Y. 2004. China Species Red List (Vol.1). Beijing: Higher Education Press.
- Wei BY. 2019. The phylogeny study of Acer L. sect. Trifoliata Pax. Jilin, China: Northeast Normal University.
- Wu ZY, Raven PH, Hong DY. 2008. Oxalidaceae through Aceraceae. Flora of China; Beijing: Science Press.p. 11.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Xia XH, Yu XD, Fu QD, Zheng YQ, Zhang CH. 2020. Complete chloroplast genome sequence of the three-flowered maple, Acertriflorum (Sapindaceae). Mitochondr DNA Part B. 5(2):1859–1860.
- Zhang DG, Xu L, Kang ZJ, Chen GX. 2011. New recorded plants from Wuling Mt. in Hunan province. J Jishou Univ (Nat Sci Ed). 32(2):78–82.
