Abstract
Ulmus parvifolia Jacq is a kind of landscape tree endemic to East Asia. In this study, the complete chloroplast genome of U. parvifolia was sequenced. The genome was 159,259 bp in length, with a large single-copy (LSC) region of 88,451 bp, a small single-copy (SSC) region of 19,598 bp, and two inverted repeat (IR) regions of 25,605 bp, each. The genome consisted of 121 genes, including 77 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The phylogenetic results indicated that Ulmus is not monophyletic and U. parvifolia constitute a well-supported clade sister to U. Sect. Ulmus. The complete chloroplast genome of U. parvifolia will provide important information for phylogenetic and evolutionary studies in Ulmaceae, as well as the other closely related families.
Ulmus L. is a genus of trees in family Ulmaceae, including 40 species and distributed in the Northern Hemisphere (Wu et al. Citation2003). Ulmus parvifolia is useful for a multitude of landscapes, as ornamental shade trees, which can survive well in climate extremes and resistant to Dutch elm diseases (Thakur and Karnosky Citation2007). Nevertheless, its genetic background and resources have not been widely studied. In this study, we reported the complete chloroplast genome sequence of U. parvifolia for future phylogenetic and genetic studies (GenBank accession number: MT604161).
A single individual of U. parvifolia was used as a sampling object from the Fudian Village (114°49′46′′ E, 31°50′12′′ N) in Xinyang City, Henan Province, China, and the voucher specimens were deposited at the Herbarium of Henan Agricultural University (voucher number ljm-20-0518). Total genomic DNA was extracted following the modified CTAB protocol (Yang et al. Citation2014), then sequenced using the Illumina HiSeq2500 platform. The generated reads were assembled by GetOrganelle v1.5.2 (Jin et al. Citation2018) with Ulmus pumila (GenBank number: KY244086) as the reference. The genome annotation was performed with CpGAVAS webserver (Liu et al. Citation2012), then the boundaries between inverted repeats were confirmed manually using Geneious v9.12 (https://www.geneious.com/). The physical map of the chloroplast genome was drawn with OGDRAW v1.3.1 (Lohse et al. Citation2007).
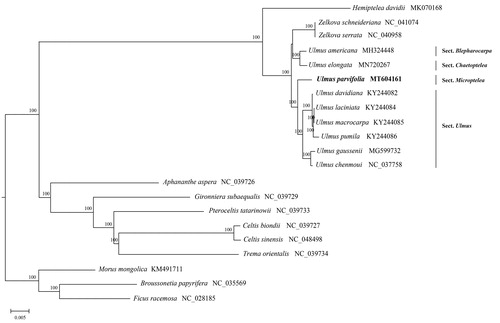
The chloroplast genome of U. parvifolia is 159,259 bp in length, and the structure was a typical quadripartite, which exhibited a large single-copy region (LSC) with 88,451 bp, a small single-copy region (SSC) with 19,598 bp, and two inverted repeat (IR) regions of 25,605 bp, each. The base compositions of U. parvifolia chloroplast genome included 31.2% A, 18.0% G, 18.7%C, 32.1% T, with an overall GC content of 35.6%. The circular genome contained 121 genes, including 77 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. Complete chloroplast genomes from Ulmaceae (17 species) and other related families (Morus mongolica, Ficus racemosa, Broussonetia papyrifera) were chosen as outgroups in the phylogenetic analysis. Sequence alignment was conducted by MAFFT v7.450 (Katoh and Standley Citation2013), and a phylogenetic tree was generated using RAxML v.8.1.2 (Stamatakis Citation2014) based on maximum-likelihood methods and the GTRGAMMA nucleotide substitution model by 1000 bootstrap replicates. Phylogenetic analysis indicated that Ulmus may be paraphyletic with Zelkova nested within it. Our results informally recognized the four subgeneric sections (Sect. Blepharocarpa, Sect. Chaetoptelea, Sect. Ulmus and Sect. Microptelea) within Ulmus, which was in accordance with previous studies (Han et al. Citation2011; Liu et al. Citation2019; Zhang et al. Citation2019). The focal species U. parvifolia constitute a clade sister to the six species of U. Sect. Ulmus. Ulmus americana from Sect. Blepharocarpa and Ulmus elongata from Sect. Chaetoptelea are more closely related to Zelkova than the rest congeneric species included (). This study was the first report on the complete chloroplast genome of U. parvifolia, which would be beneficial to potential studies on phylogenetics of the genus and related group in Ulmaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov/, reference number MT604161.
Additional information
Funding
References
- Han X, Zhi YB, Zhang BW, Zhou ZZ, He JQ, Wang ZL, Li HL, Li JM. 2011. The phylogenetic relationships between Ulmus gaussenii Cheng and its close relatives in genus Ulmus based on the ITS and atpB-rbcL. J Anhui Univ. 35 (6):98–107.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv. 4:256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13(1):715.
- Liu H, Zhang Y, Zhang X, Geng M, Li M. 2019. The first complete chloroplast genome of Zelkova schneideriana Hand.-Mazz. (Ulmaceae). Mitochondrial DNA Part B. 4(1):378–379.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Thakur RC, Karnosky DF. 2007. Micropropagation and germplasm conservation of Central Park Splendor Chinese elm (Ulmus parvifolia Jacq. ‘A/Ross Central Park’) trees. Plant Cell Rep. 26(8):1171–1177.
- Wu ZY, Raven PH, Hong DY. 2003. Ulmaceae through Basellaceae. Flora of China. 88(1):83–272.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031.
- Zhang Q, Zhang H, Li Q, Bai R, Ning E, Cai X. 2019. Characterization of the complete chloroplast genome sequence of an endangered elm species, Ulmus gaussenii (Ulmaceae). Conservation Genet Resour. 11(1):71–74.