Abstract
Freshwater mussels are a critically imperiled group of mollusks that play key ecological roles and provide important services to humans. The Ambleminae is the only subfamily of these mussels, endemic to North America. Complete mitogenomes have only been sequenced for two of five tribes of the subfamily. Pleurobema oviforme, Amblema plicata, and Popenaias popeii each belong to tribes Pleurobemini, Amblemini, and Popenaidini, respectively, and have not had published mitogenomes. Thus, this study aims to present the complete mitogenomes for these species, to provide a phylogeny of the Ambleminae and confirm the gene arrangements with representation from each of its tribes. The newly sequenced mitogenomes range from 15,852 to 15,993 nucleotides, are composed of 13 PCGs, 22 tRNAs, and two rRNAs and all share the same (UF1) gene order.
Freshwater mussels (Bivalvia: Unionida) are important members of aquatic ecosystems and are among the most threatened faunistic groups worldwide. Both North and Central America are among the regions with higher freshwater mussel diversity (Lopes-Lima et al. Citation2018). However, these taxa have suffered a dramatic decline and a high number of extinctions in these regions (Haag and Williams Citation2014), emphasizing the need for understanding their evolutionary relationships to guide future conservation measures (Levine et al. Citation2012). To date, three subfamilies are recognized within the Unionidae in the North and Central America: Gonideinae Ortmann (1916), Unioninae Rafinesque (1820), and Ambleminae Rafinesque (1820). The Ambleminae is the only subfamily endemic in this region. Several studies have been published to characterize the phylogenetic relationships within the Ambleminae in North America, although genera that occur in Central America were rarely included. The most recent classification separates the amblemines into five tribes: Lampsilini, Popenaiadini, Amblemini, Pleurobemini, and Quadrulini (Pfeiffer et al. Citation2019). Complete mitogenomes are only available for two of these tribes, Lampsilini with eight, Quadrulini with two, Pleurobemini with only one partial mitogenome and Popenaiadini and Amblemini with no available complete mitogenome. Freshwater mussels present different gene orders that might be useful for phylogenetic inference. Within Unionidae, the gene order UF1 is shared among all members of the Ambleminae and Unioninae subfamilies, while UF2 and UF3 occur in members of Gonideinae (Lopes-Lima et al. Citation2017).
Here, we present the complete mitogenomes of Amblema plicata (Say, 1817), Pleurobema oviforme (Conrad, 1834), and Popenaias popeii (Lea, 1857) and their evolutionary relationships. The latter two have a high conservation concern being considered as threatened by the IUCN Red List (https://www.iucnredlist.org/).
Pleurobema oviforme was collected from the Little Tennessee River, Tennessee 35.325820 longitude 83.523930 latitude and deposited in North Carolina Museum of Natural Sciences USA (voucher: NCSM30708). Popenaias popeii was collected in Rio Grande River, Texas 99.49890 longitude, 27.49921 latitude, and deposited in the Great Lakes Center Invertebrate Collection (BSGLC) (SUNY Buffalo State College, Buffalo, NY, USA) (voucher: 067TS). Amblema plicata was collected in the San Marcos River, TX, 29.588390 latitude 97.586110 longitude, deposited in the BSGLC (voucher: 061TS). The collection in Texas was carried out with an appropriate Scientific Research Permit SPR-0503-300 issued by the Texas Parks and Wildlife Department. Genomic DNA was extracted using standard protocols and thereafter sequenced using MiSeq Illumina runs. Resulting reads were trimmed in Trimmomatic (Bolger et al. Citation2014). Each mitogenome was assembled using MITObim (Hahn et al. Citation2013), and annotated in MITOS (Bernt et al. Citation2013). Additional sequences of all Ambleminae species were downloaded from GenBank together with two margaritiferid species as outgroup. Each individual protein-coding gene and rRNA gene were aligned and trimmed using GUIDANCE2 (Sela et al. Citation2015) with the MAFFT algorithm (Katoh and Standley Citation2013). The resulting trimmed alignments of the individual genes were concatenated. The phylogenetic relationships were inferred by maximum likelihood (ML) using IQTree version 1.6.10 (Nguyen et al. Citation2015) with 20,000 ultrafast-bootstraps and Bayesian inference (BI) using MrBayes version 3.2.7a (Ronquist et al. Citation2012) with two independent runs of 107 generations and a sampling frequency from every 1,000 trees. The best partition models were selected under the software PartitionFinder version 2 2.2.1 (Lanfear et al. Citation2016) for both IQ-Tree and MrBayes. These were run on XSEDE through the CIPRES Science Gateway (Miller et al. Citation2010). Complete mitogenomes of A. plicata, P. oviforme, and P. popeii were submitted to GenBank: MT648774, MT648775, and MT648776. Each contained 15,946, 15,852, and 15,993 bps length, respectively, and are composed by 13 PCGs, 2 rRNAs, and 22 tRNAs.
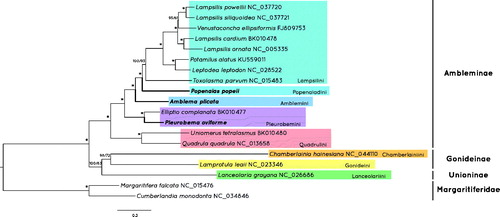
The resulting ML and BI trees share the same topology (), rooted with the two margaritiferid species. An initial split occurs between the Ambleminae and Gonideinae + Unioninae. The Ambleminae is further separated between Quadrilini and the rest of the tribes. The new mitogenome of P. oviforme clusters with E. complanata in a well-supported clade. Both A. plicata (Amblemini) and P. popeii (Popenaiadini) represent the first available mitogenomes within their tribes and therefore do not show any close phylogenetic relationship. This tree confirms the same phylogeny of the most recent tribal classification within Ambleminae (Pfeiffer et al. Citation2019). Finally, all Ambleminae mitogenomes (including the newly sequenced) share the same gene order, i.e. UF1.
Figure 1. Bayesian Inference phylogenetic tree of the concatenated protein coding and rRNA genes of the available mitogenomes of Ambleminae and two outgroup taxa. Values in branches represent the Bayesian posterior probabilities percentage obtained by MrBayes and the bootstrap support values from IQ‐Tree. Values above 95% for both are replaced with an *.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data produced in this study are available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference numbers MT648774-MT648776 or from the corresponding author.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Haag WR, Williams JD. 2014. Biodiversity on the brink: an assessment of conservation strategies for North American freshwater mussels. Hydrobiologia. 735(1):45–60.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acid Res. 41(13):e129–e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. Partition finder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34:772–773.
- Levine TD, Lang BK, Berg DJ. 2012. Physiological and exological hosts of Popenaias popeii (Bivalvia: Unionidae): laboratory studies identify more hosts than field studies. Freshwater Biol. 57(9):1854–1864.
- Lopes-Lima M, Burlakova LE, Karatayev AY, Mehler K, Seddon M, Sousa R. 2018. Conservation of freshwater bivalves at the global scale: diversity, threats and research needs. Hydrobiologia. 810(1):1–14.
- Lopes-Lima M, Fonseca MM, Aldridge DC, Bogan AE, Gan HM, Ghamizi M, Sousa R, Teixeira A, Varandas S, Zanatta D, et al. 2017. The first Margaritiferidae male (M-type) mitogenome: mitochondrial gene order as a potential character for determining higher-order phylogeny within Unionida (Bivalvia). J Molluscan Stud. 83(2):249–252.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for interference for large phylogenetic trees. Proceedings of the gateway computing environments workshop (GCE), IEEE, 14 Nov. 2010, New Orleans (LA); p. 1–8.
- Nguyen LT, Schmidt HA, Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Pfeiffer JM, Atkinson CL, Sharpe AE, Capps KA, Emery KF, Page LM. 2019. Phylogeny of Mesoamerican freshwater mussels and a revised tribe-level classification of the Ambleminae. Zool Scr. 48(1):106–112.
- Ronquist F, Teslenko M, Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Sela I, Ashkenazy H, Katoh K, Pupko T. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43(W1):W7–W14.
